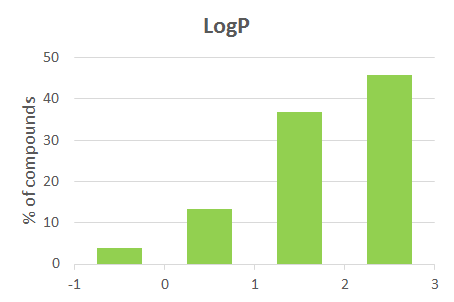What makes the fragment a specific “key” to the binding site is its shape. Fragment libraries mostly consist of rod-like molecules, with spherical ones highly diluted in this number. Disc- and sphere-shaped molecules are more conformationally rigid which contributes to the unambiguity of their interaction with the target. The shape of the molecule can be predicted from the Plane of Best Fit and Principal Moments of Inertia descriptors.
For Chemspace 3D-Shaped fragments we selected only the compounds that have PBF≥0.6 and NPRsum≥1.1 and, at the same time, their shape determined from the PMI1-PMI2-PMI3 triangle diagram is not rod-like. These fragments have mostly, but not necessarily, high fsp3 value.
For any questions contact us at [email protected]
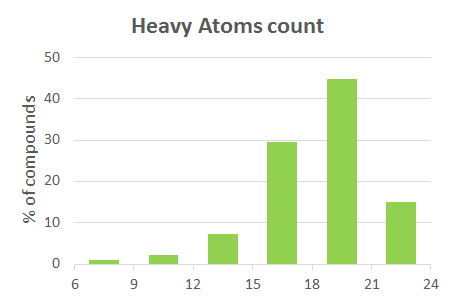
| PhysChem Profile | |||
|---|---|---|---|
| Min | Max | Median | |
| 6 | Heavy Atoms count | 22 | 18 |
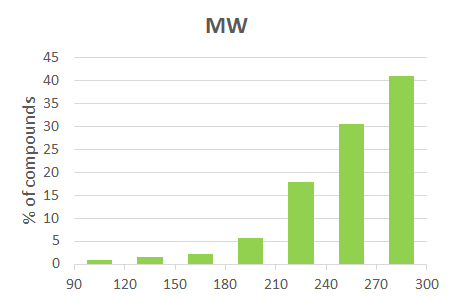
| 85 | MW | 300 | 257 |
| -1.2 | SlogP | 3 | 1.7 |
| 1 | HBA count | 3 | 3 |
| 0 | HBD count | 3 | 1 |
| 0 | RB count | 3 | 2 |
| 0.1 | fsp3 | 1 | 0.5 |
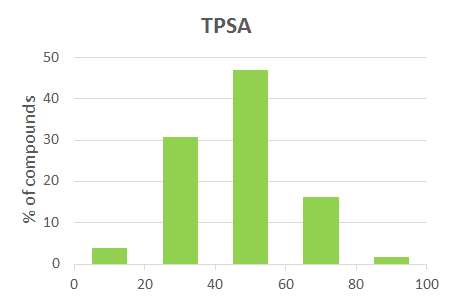
| 12 | TPSA | 92 | 49 |
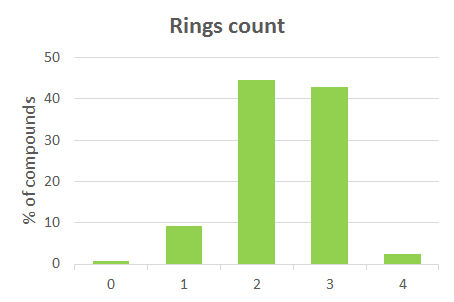
| 0 | Rings count | 4 | 2 |