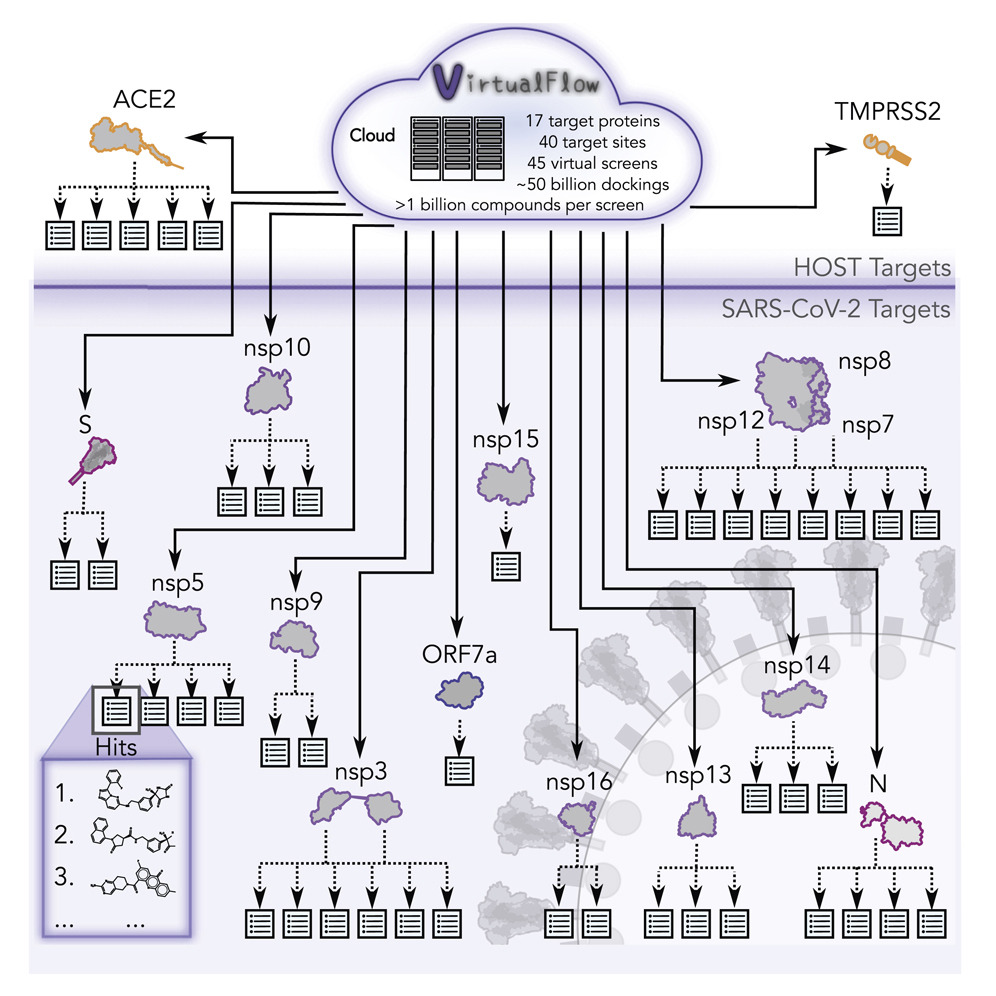
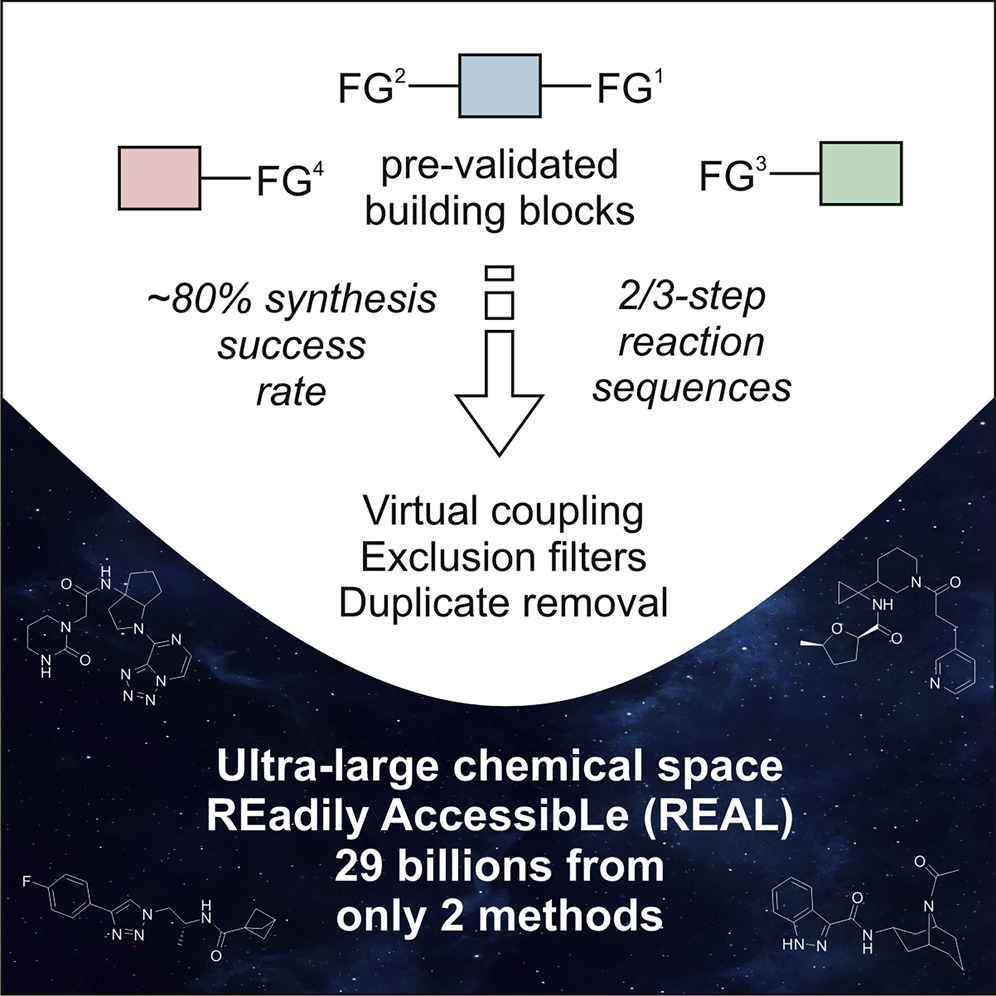
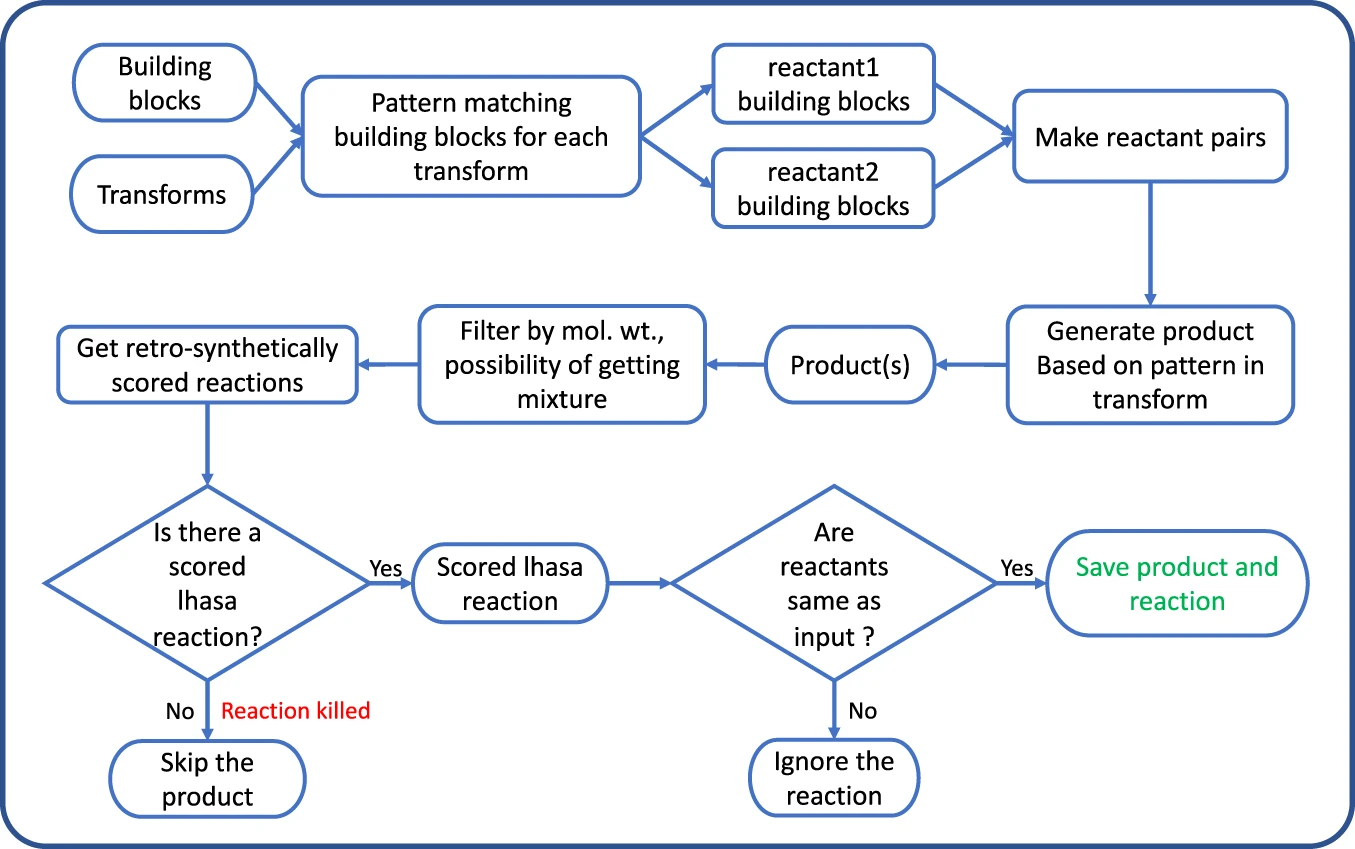
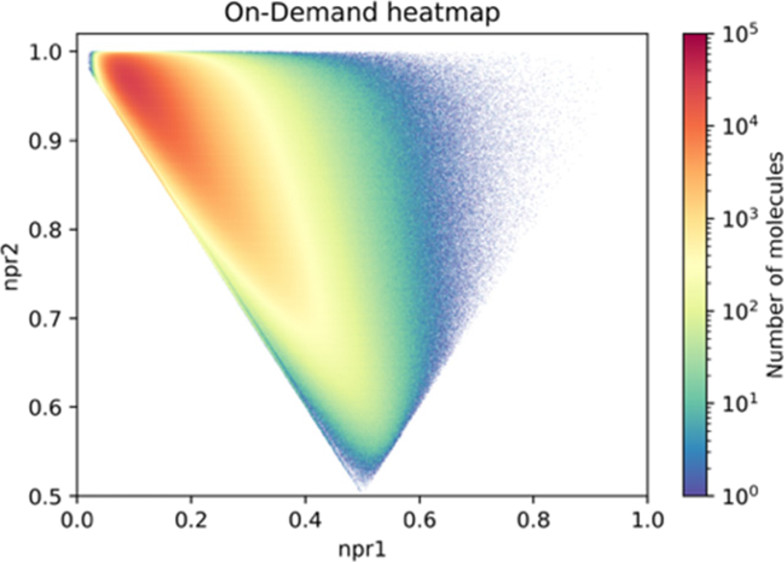
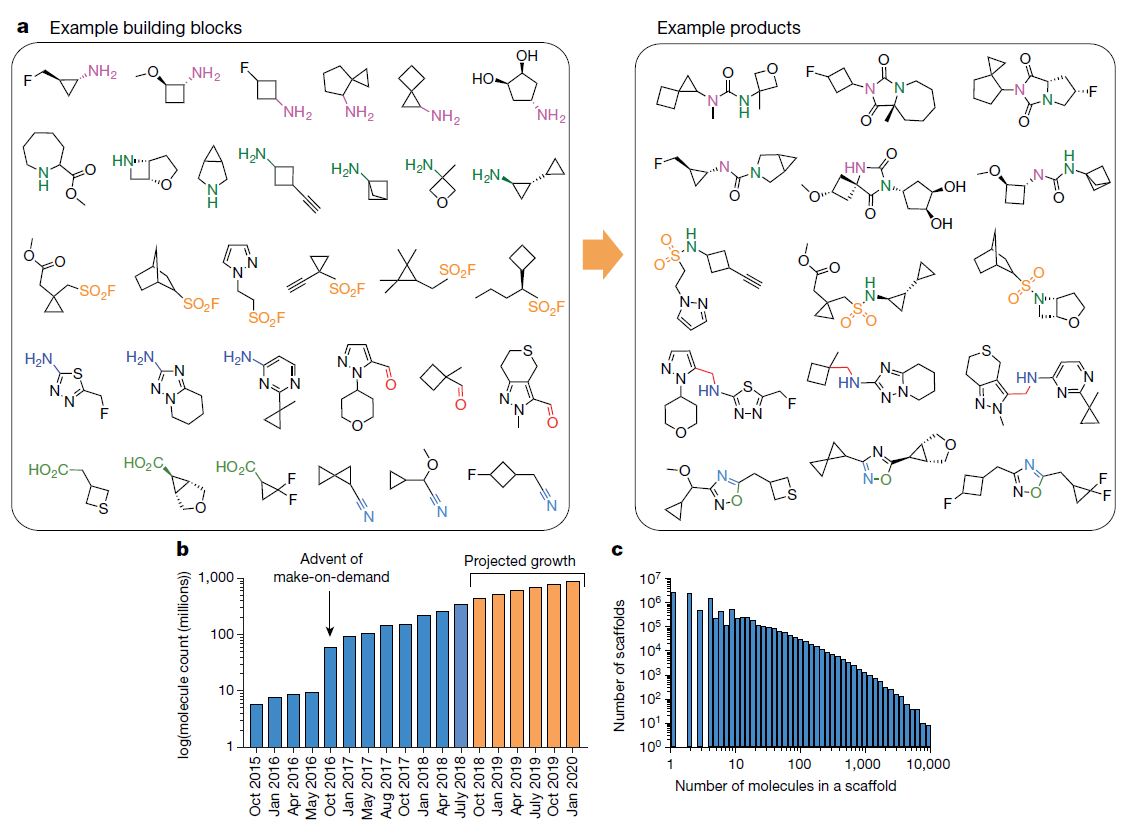
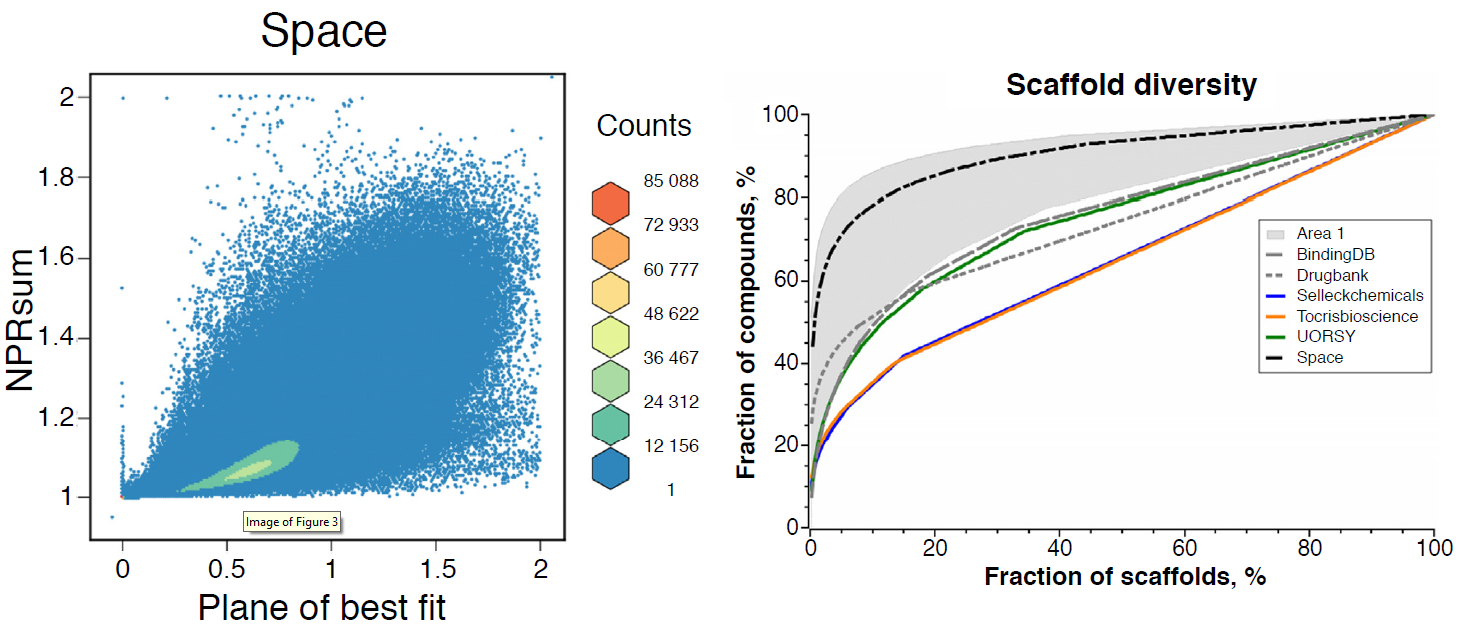
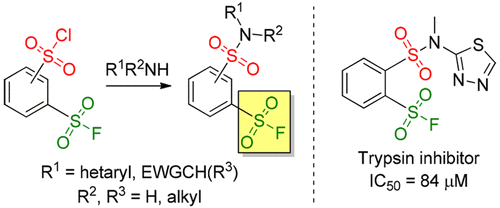
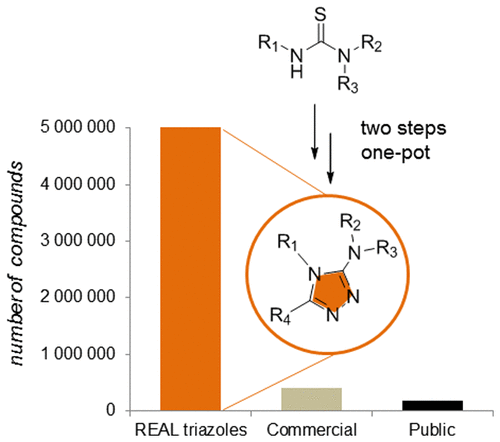
-
1. Serendipitous Discovery of an Allosteric Inhibitor Binding Groove in the Proline Biosynthetic Enzyme Pyrroline-5-Carboxylate Reductase 1 (PYCR1)
Kaylen R Meeks, Caitlin J Mattingly, Jay C Nix, Oleksii Chuk, Mykola V Protopopov, Olga O Tarkhanova, John J Tanner


-
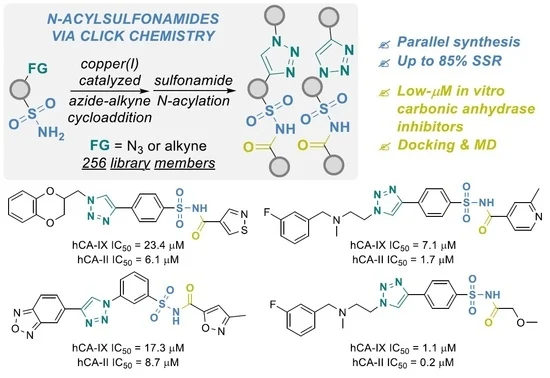
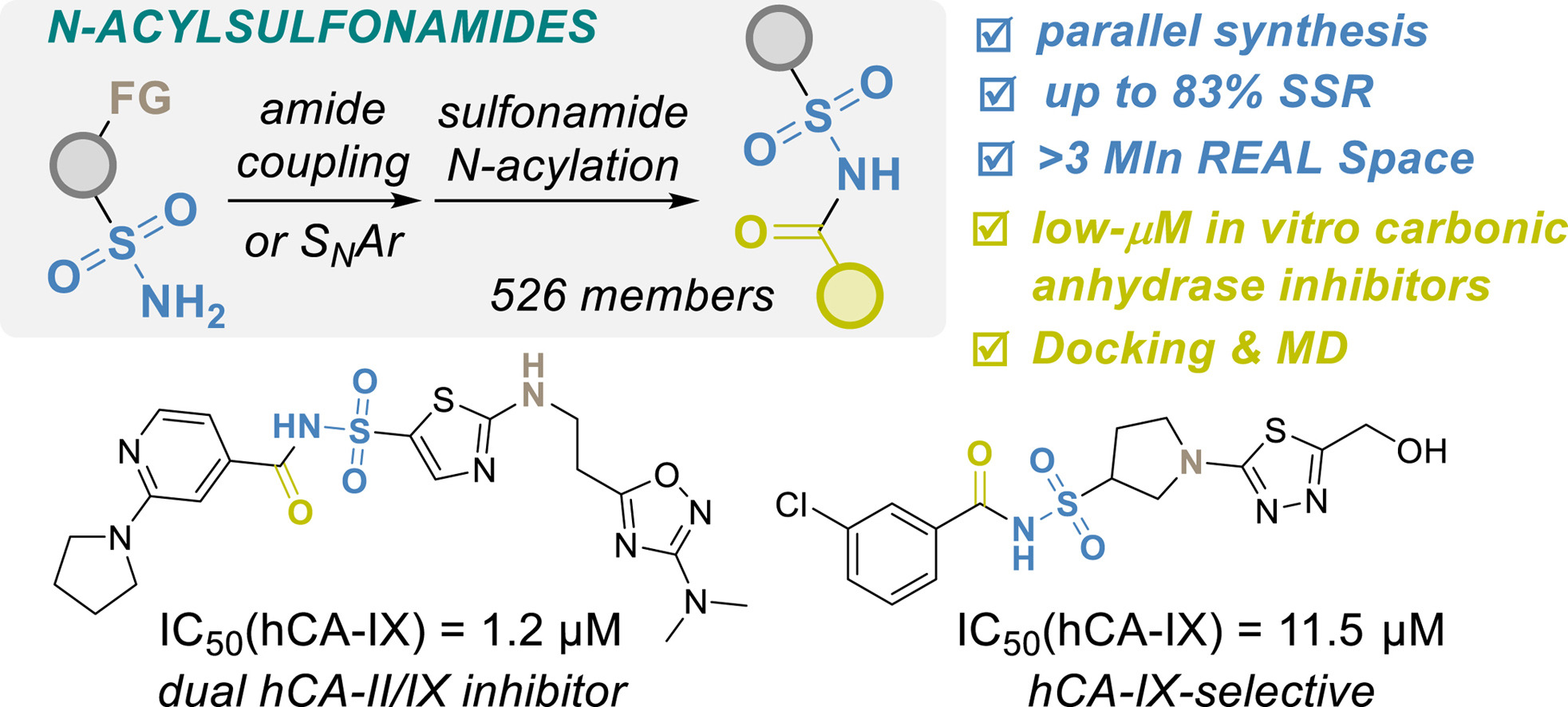
2. Click Chemistry-Enabled Parallel Synthesis of N-Acyl Sulfonamides and Their Evaluation as Carbonic Anhydrase Inhibitors
Oleksii V. Gavrylenko, Bohdan V. Vashchenko, Vasyl Naumchyk, Bohdan S. Sosunovych, Oleksii Chuk, Oleksii Hrabovskyi, Olga Kuchuk, Alla Pogribna, Sergiy O. Nikitin, Anzhelika I. Konovets, Volodymyr S. Brovarets, Sergey A. Zozulya, Dmytro S. Radchenko, Oleksandr O. Grygorenko, Yurii S. Moroz

-
3. Expanding Chemical Space of N-Acyl Sulfonamides for Carbonic Anhydrase Inhibitor Discovery
Oleksii V. Gavrylenko, Bohdan V. Vashchenko, Vasyl Naumchyk, Oleksii Chuk, Olga Kuchuk, Alla Pogribna, Anzhelika I. Konovets, Volodymyr S. Brovarets, Sergey A. Zozulya, Dmytro S. Radchenko, Oleksandr O. Grygorenko, Yurii S. Moroz

-
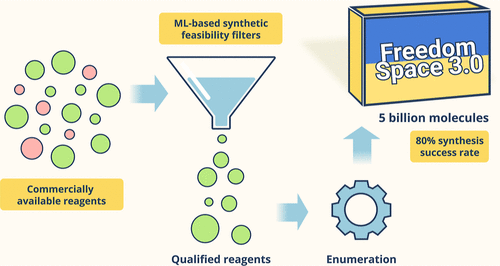
4. Freedom Space 3.0: ML-Assisted Selection of Synthetically Accessible Small Molecules
Anna Kapeliukha, Serhii Hlotov, Mykola Protopopov, Igor Dzyuba, Maryna Vasylchuk, Dmitriy M. Panov, Olga O. Tarkhanova, Yurii S. Moroz

-
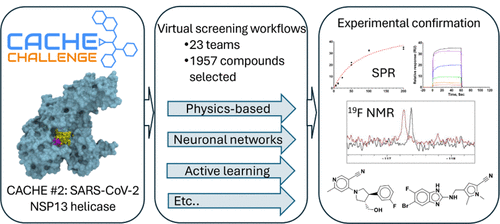
5. CACHE Challenge #2: Targeting the RNA Site of the SARS-CoV-2 Helicase Nsp13
Oleksandra Herasymenko, Madhushika Silva, Abd Al-Aziz A. Abu-Saleh, Ayaz Ahmad, Jesus Alvarado-Huayhuaz, Oscar E. A. Arce, Roly J. Armstrong, Cheryl Arrowsmith, Kelly E. Bachta, Hartmut Beck, Denes Berta, Mateusz K. Bieniek, Vincent Blay, Albina Bolotokova, Philip E. Bourne, Marko Breznik, Peter J. Brown, Aaron D. G. Campbell, Emanuele Carosati, Irene Chau, Daniel J. Cole, Ben Cree, Wim Dehaen, Katrin Denzinger, Karina dos Santos Machado, Ian Dunn, Prasannavenkatesh Durai, Kristina Edfeldt, Aled Edwards, Darren Fayne, Daniel Felfoldi, Kallie Friston, Pegah Ghiabi, Elisa Gibson, Judith Günther, Anders Gunnarsson, Alexander Hillisch, Douglas R. Houston, Jan Halborg Jensen, Rachel J. Harding, Kate S. Harris, Laurent Hoffer, Anders Hogner, Joshua T. Horton, Scott Houliston, Judd F. Hultquist, Ashley Hutchinson, John J. Irwin, Marko Jukič, Shubhangi Kandwal, Andrea Karlova, Vittorio L. Katis, Ryan P. Kich, Dmitri Kireev, David Koes, Nicole L. Inniss, Uta Lessel, Sijie Liu, Peter Loppnau, Wei LuSam Martino, Miles McGibbon, Jens Meiler, Akhila Mettu, Sam Money-Kyrle, Rocco Moretti, Yurii S. Moroz, Charuvaka Muvva, Joseph A. Newman, Leon Obendorf, Brooks Paige, Amit Pandit, Keunwan Park, Sumera Perveen, Rachael Pirie, Gennady Poda, Mykola Protopopov, Vera Pütter, Federico Ricci, Natalie J. Roper, Edina Rosta, Margarita Rzhetskaya, Yogesh Sabnis, Karla J. F. Satchell, Frederico Schmitt Kremer, Thomas Scott, Almagul Seitova, Casper Steinmann, Valerij Talagayev, Olga O. Tarkhanova, Natalie J. Tatum, Dakota Treleaven, Adriano Velasque Werhli, W. Patrick Walters, Xiaowen WangJude Wells, Geoffrey Wells, Yvonne Westermaier, Gerhard Wolber, Lars Wortmann, Jixian Zhang, Zheng Zhao, Shuangjia Zheng, Matthieu Schapira

-
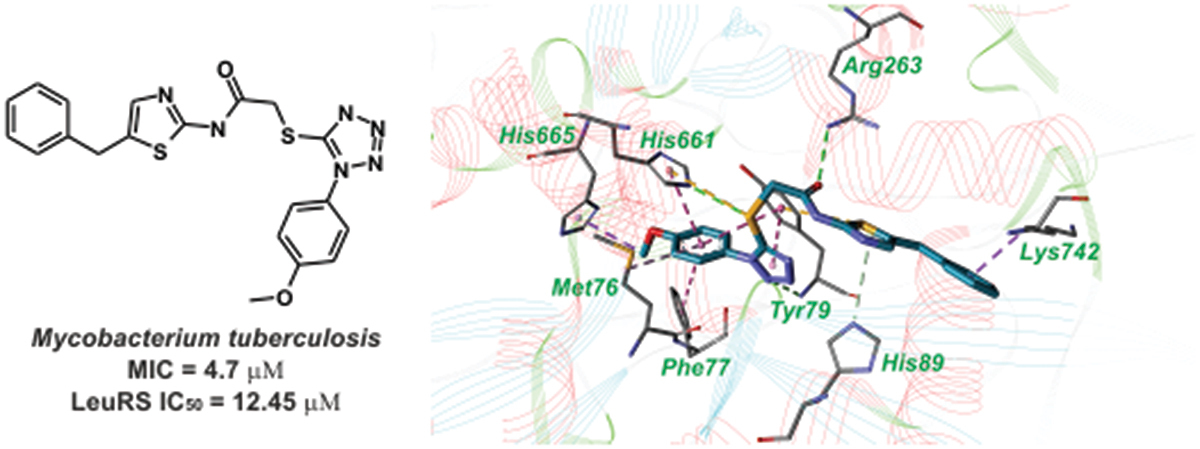
6. Identification of novel Mycobacterium tuberculosis leucyl-tRNA synthetase inhibitors with antibacterial activity
Galyna P. Volynets, Olga I. Gudzera, Segiy S. Lukashov, Oksana B. Gorbatiuk, Mariia O. Usenko, Tetiana P. Ruban, Mykola V. Protopopov, Segiy S. Tarnavskiy, Igor M. Kotey, Volodymyr G. Bdzhola, Andrii O. Prykhod’ko, Lubov L. Lukash, Sergiy M. Yarmoluk, Michael A. Tukalo

-
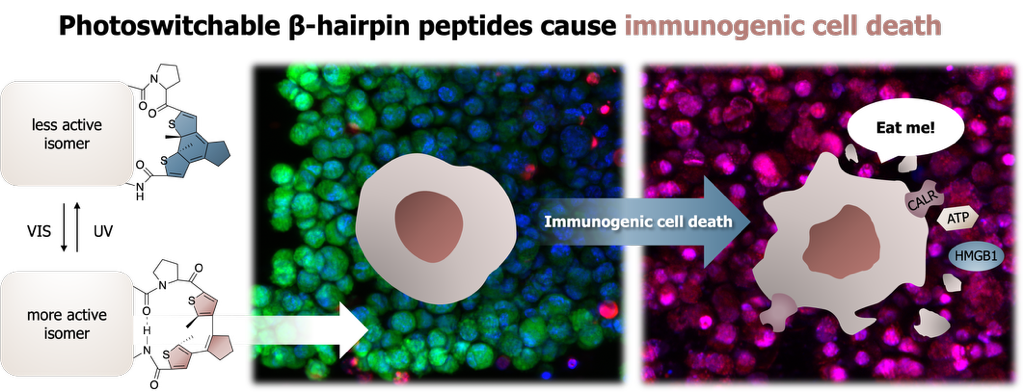
7. In vitro evaluation of the immunogenic potential of gramicidin S and its photocontrolled analogues
Kateryna Horbatok, Iryna Semchuk, Oleksandr Horbach, Natalia Khranovska, Viktoriia Kosach, Petro Borysko, Serhii Koniev, Anne S. Ulrich, Sergii Afonin, Igor V. Komarov

-
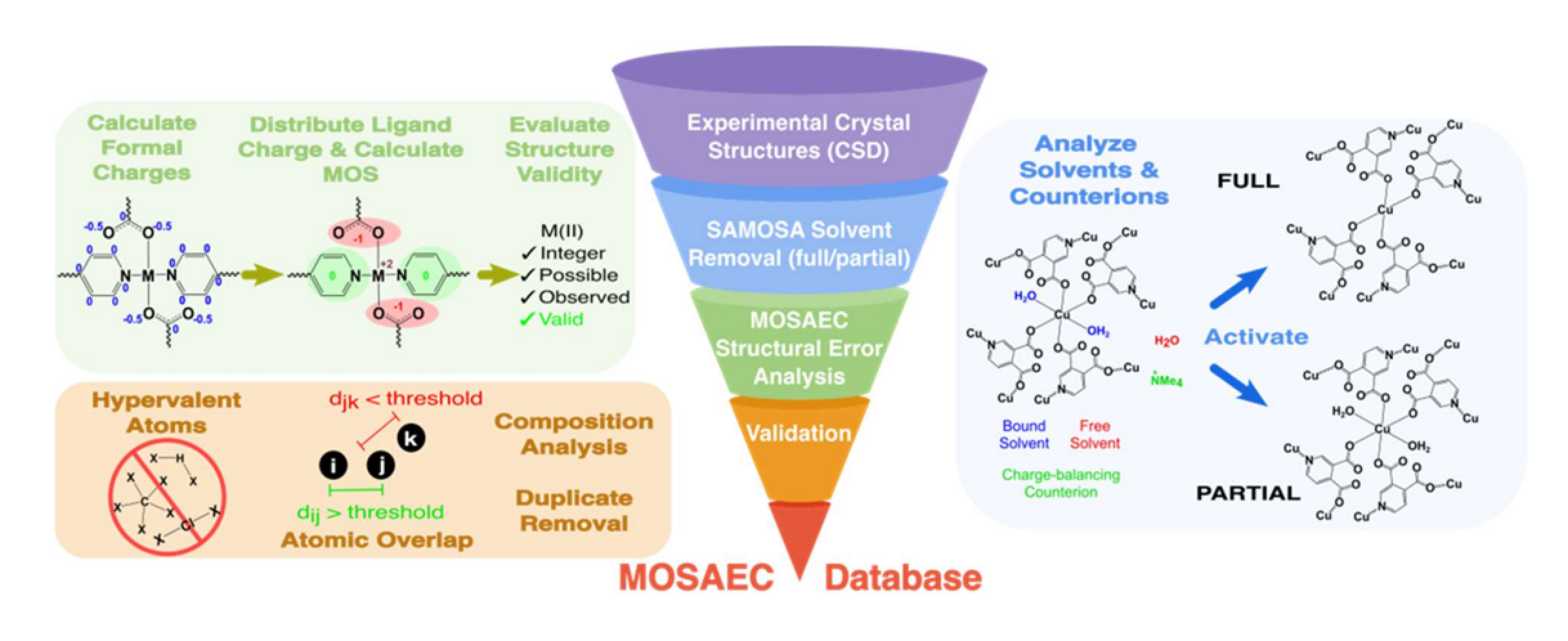
8. MOSAEC-DB: a comprehensive database of experimental metal–organic frameworks with verified chemical accuracy suitable for molecular simulations
Marco Gibaldi, Anna Kapeliukha, Andrew White, Jun Luo, Robert Alex Mayo, Jake Burner and Tom K. Woo

-
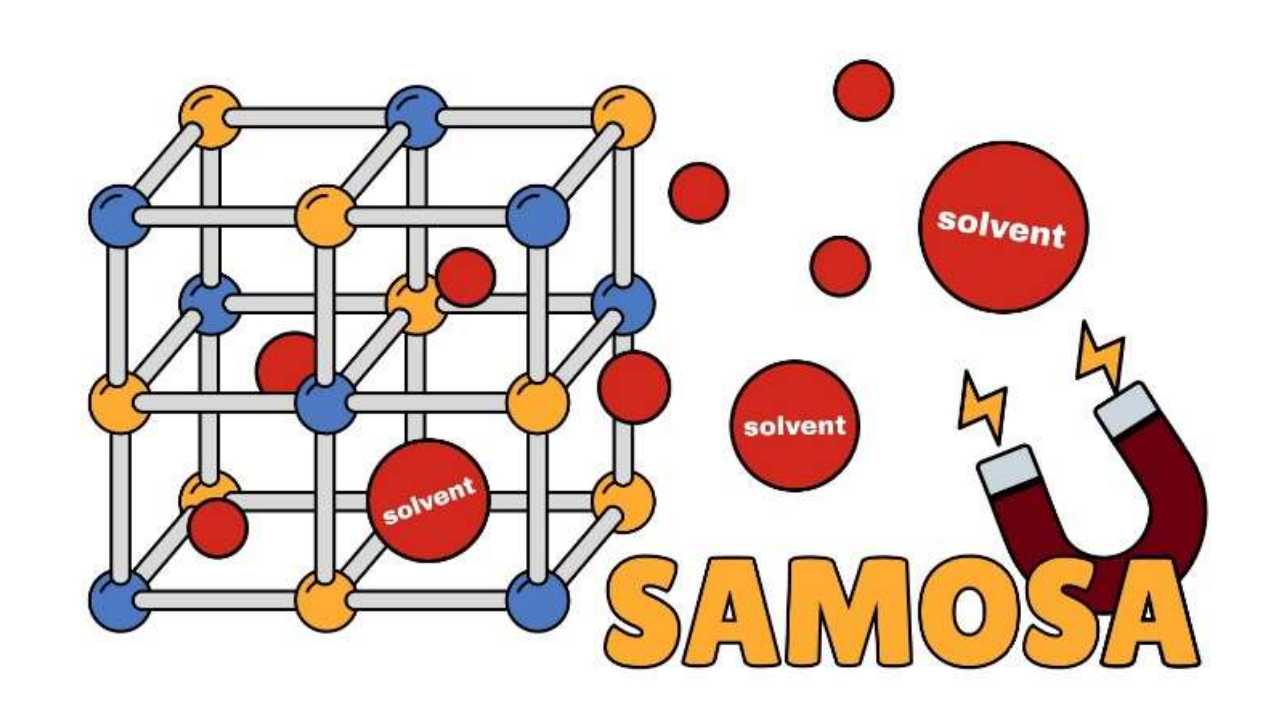
9. Incorporation of Ligand Charge and Metal Oxidation State Considerations into the Computational Solvent Removal and Activation of Experimental Crystal Structures Preceding Molecular Simulation
Marco Gibaldi, Anna Kapeliukha, Andrew White, Tom K. Woo

-
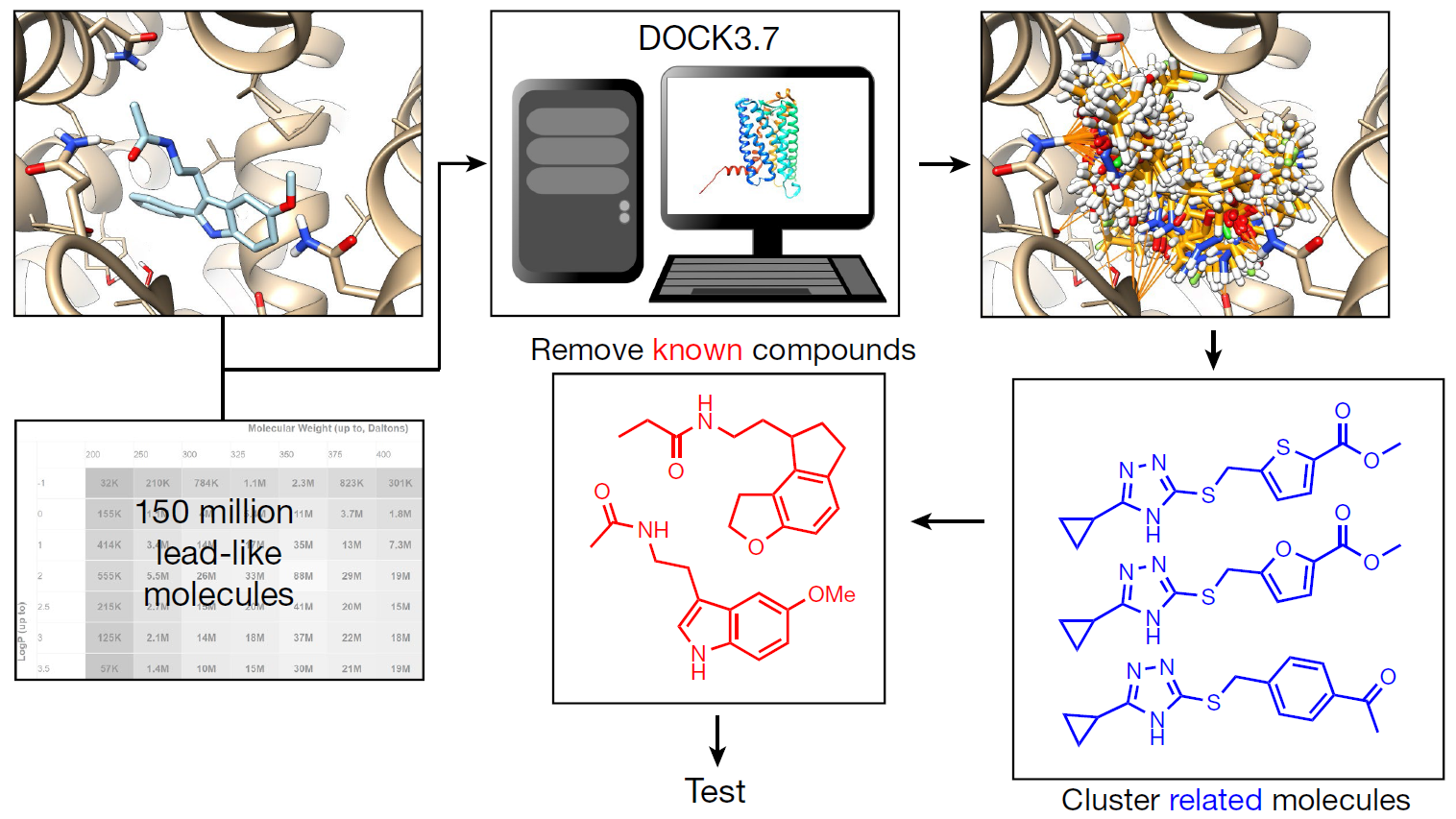
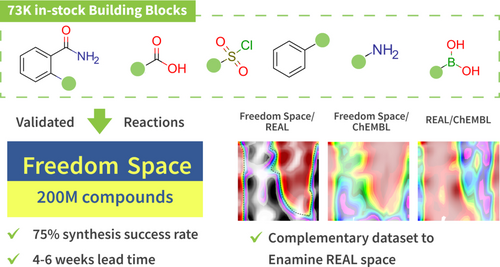
10. The freedom space – a new set of commercially available molecules for hit discovery
Mykola V. Protopopov, Valentyna V. Tararina, Fanny Bonachera, Igor M.Dzyuba, Anna Kapeliukha, Serhii Hlotov, Oleksii Chuk, Gilles Marcou, Olga Klimchuk, Dragos Horvath, Erik Yeghyan, Olena Savych, Olga O. Tarkhanova, Alexandre Varnek, Yurii S. Moroz

-
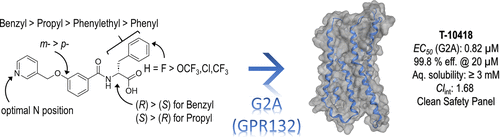
11. Development of a Potent and Selective G2A (GPR132) Agonist
Victor Hernandez-Olmos, Jan Heering, Beatrice Marinescu, Tina Schermeng, Vladimir V. Ivanov, Yurii S. Moroz, Sheila Nevermann, Marius Mathes, Johanna H. M. Ehrler, Mohamad Wessam Alnouri, Markus Wolf, Alicia S. Haydo, Tessa Schmachtel, Andrea Zaliani, Georg Höfner, Astrid Kaiser, Manfred Schubert-Zsilavecz, Annette G. Beck-Sickinger, Stefan Offermanns, Philipp Gribbon, Michael A. Rieger, Daniel Merk, Marco Sisignano, Dieter Steinhilber, and Ewgenij Proschak

-
12. Structure-based discovery of CFTR potentiators and inhibitors
Fangyu Liu, Anat Levit Kaplan, Jesper Levring, Jürgen Einsiedel, Stephanie Tiedt, Katharina Distler, Natalie S. Omattage, Ivan S. Kondratov,
Yurii S. Moroz, Harlan L. Pietz, John J. Irwin, Peter Gmeiner, Brian K. Shoichet, Jue Chen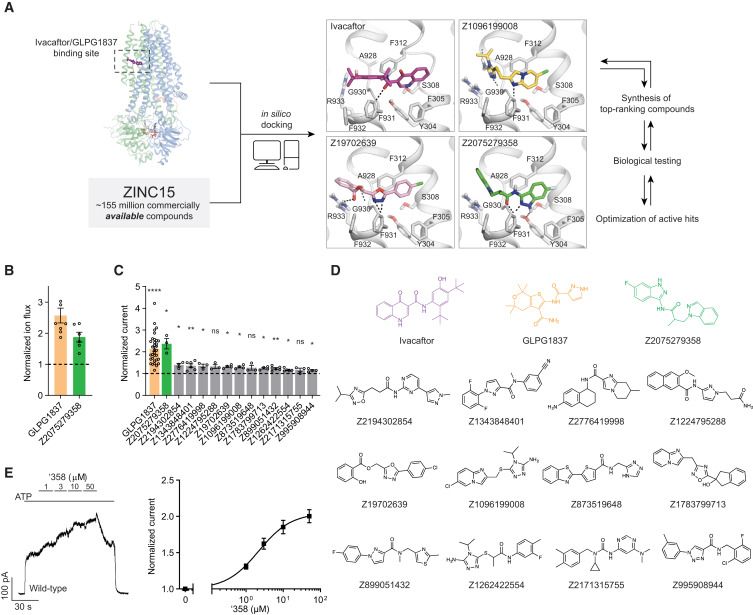

-
13. AlphaFold2 structures guide prospective ligand discovery
Jiankun Lyu, Nicholas Kapolka, Ryan Gumpper, Assaf Alon, Liang Wang, Manish K. Jain, Ximena Barros-Álvarez, Kensuke Sakamoto, Yoojoong Kim, Jeffrey DiBerto,
Kuglae Kim, Isabella S. Glenn, Tia A. Tummino, Sijie Huang, John J. Irwin, Olga O. Tarkhanova, Yurii Moroz, Georgios Skiniotis, Andrew C. Kruse, Brian K. Shoichet,
Bryan L. Roth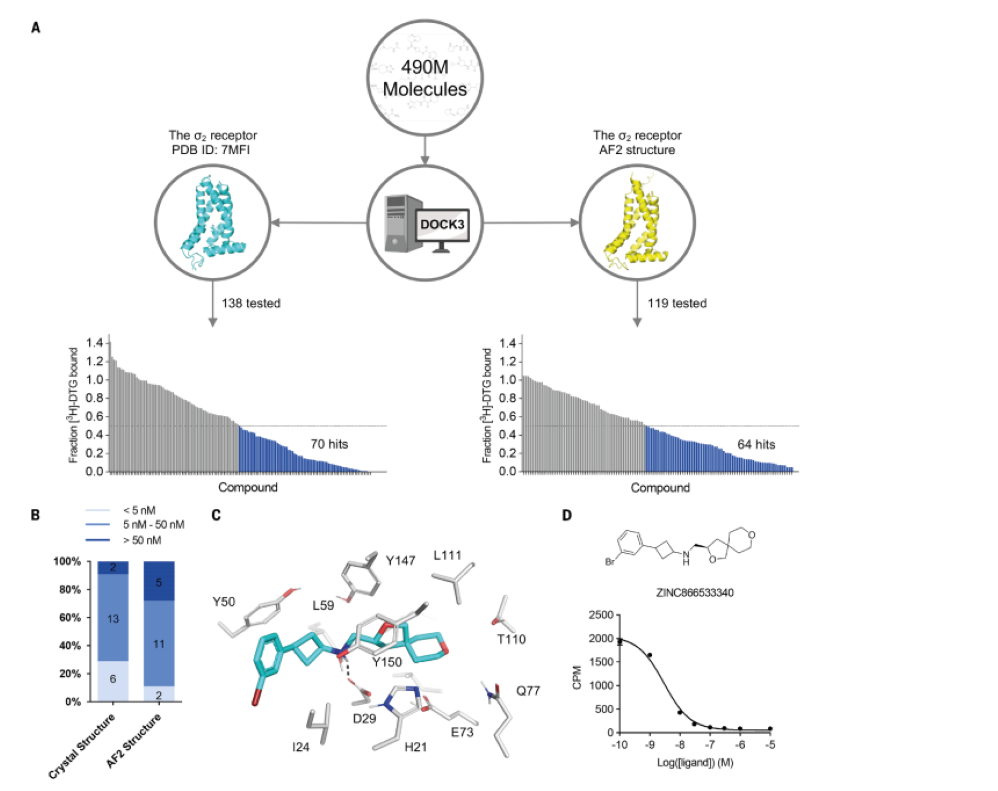

-
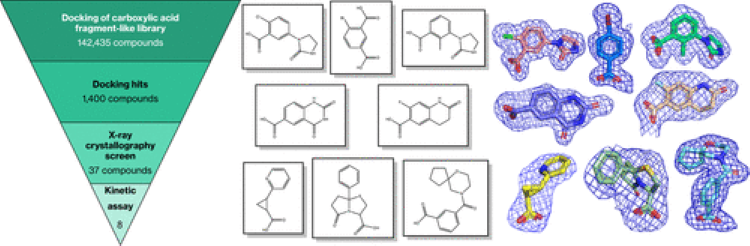
14. Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography
Kaylen R. Meeks, Juan Ji, Mykola V. Protopopov, Olga O. Tarkhanova, Yurii S. Moroz, and John J. Tanner

-
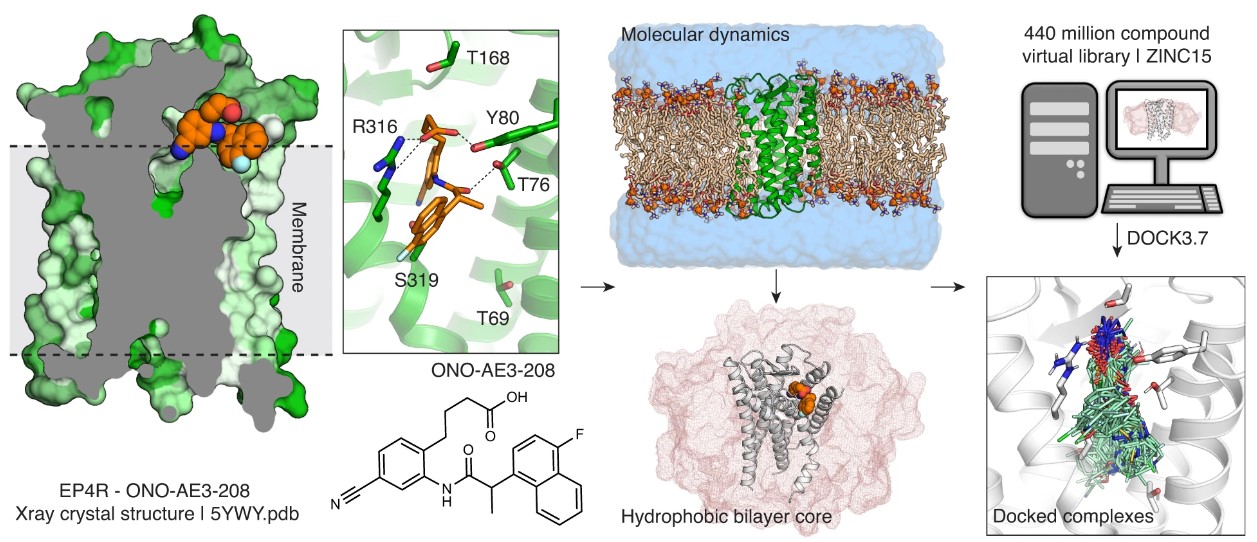
15. Docking for EP4R antagonists active against inflammatory pain
S. Gahbauer, C. DeLeon, J. M. Braz, V. Craik, H. J. Kang, X. Wan, X.-P. Huang, C. B. Billesbølle, Y. Liu, T. Che, I. Deshpande, M. Jewell, E. A. Fink, I. S. Kondratov, Y. S. Moroz, J. J. Irwin, A. I. Basbaum, B. L. Roth, B. K. Shoichet

-
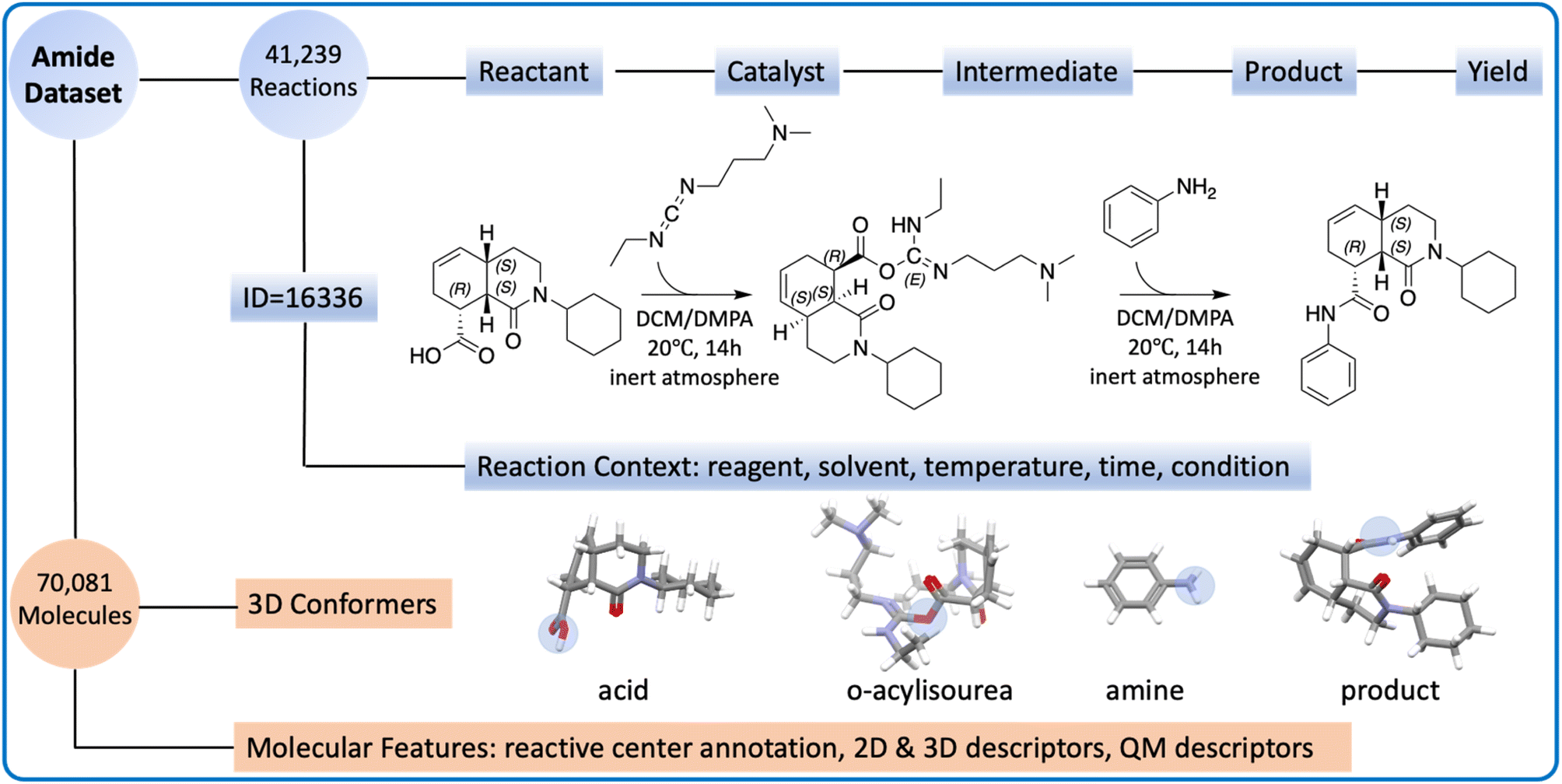
16. The challenge of balancing model sensitivity and robustness in predicting yields: a benchmarking study of amide coupling reactions
Z. Liu, Y. S. Moroz, O. Isayev

-
17. AI-Powered Virtual Screening of Large Compound Libraries Leads to the Discovery of Novel Inhibitors of Sirtuin-1
A. Gryniukova, F. Kaiser, I. Myziuk, D. Alieksieieva, C. Leberecht, P. P. Heym, O. O. Tarkhanova, Y. S. Moroz, P. Borysko, V. J. Haupt

-
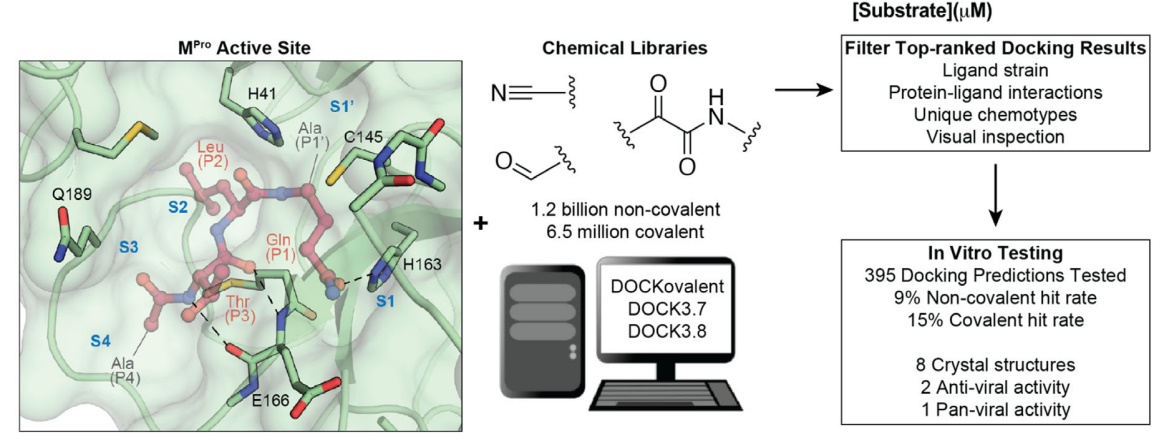
18. Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors
E. A. Fink, C. Bardine, S. Gahbauer, I. Singh, T. C. Detomasi, K. White, S. Gu, X. Wan, J. Chen, B. Ary, I. Glenn, J. O’Connell, H. O’Donnell, P. Fajtová, J. Lyu, S. Vigneron, N. J. Young, I. S. Kondratov, A. Alisoltani, L. M. Simons, R. Lorenzo‐Redondo, E. A. Ozer, J. F. Hultquist, A. J. O’Donoghue, Y. S. Moroz, J. Taunton, A. R. Renslo, J. J. Irwin, A. García‐Sastre, B. K. Shoichet, C. S. Craik

-
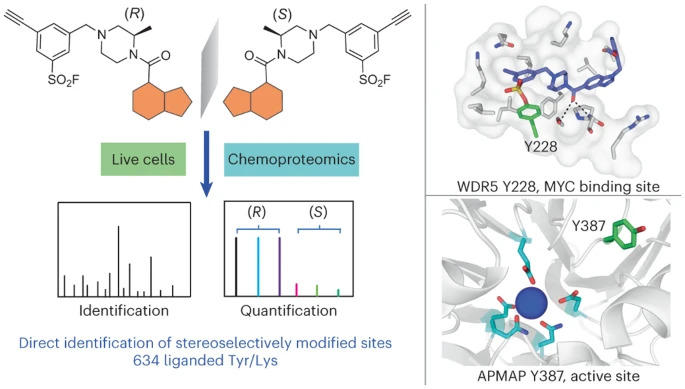
19. Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes
Y. Chen, G. B. Craven, R. A. Kamber, A. Cuesta, S. Zhersh, Y. S. Moroz, M. C. Bassik, J. Taunton

-
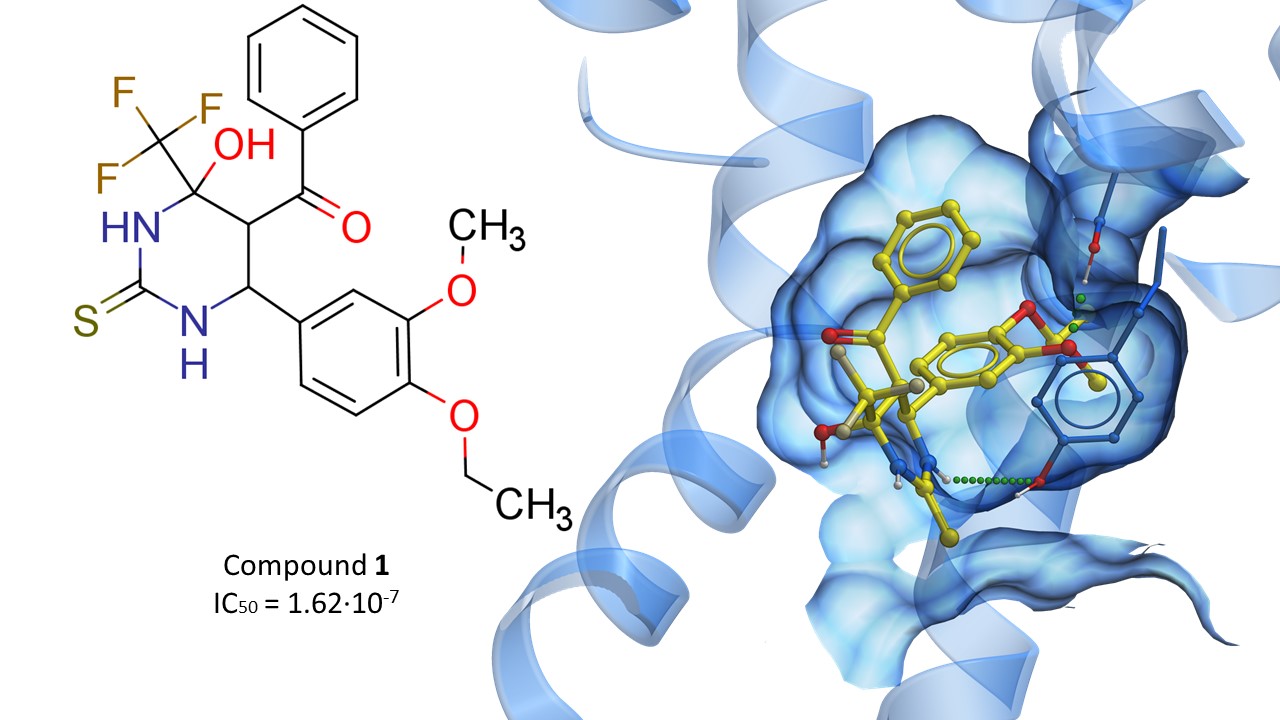
20. Computer-Aided Design Of Muscarinic Acetylcholine Receptor M3 Inhibitors: Promising Compounds Among Trifluoromethyl Containing Hexahydropyrimidinones/Thiones
A. Nyporko, O. Tsymbalyuk, I. Voiteshenko, S. Starosyla, M. Protopopov, V. Bdzhola

-
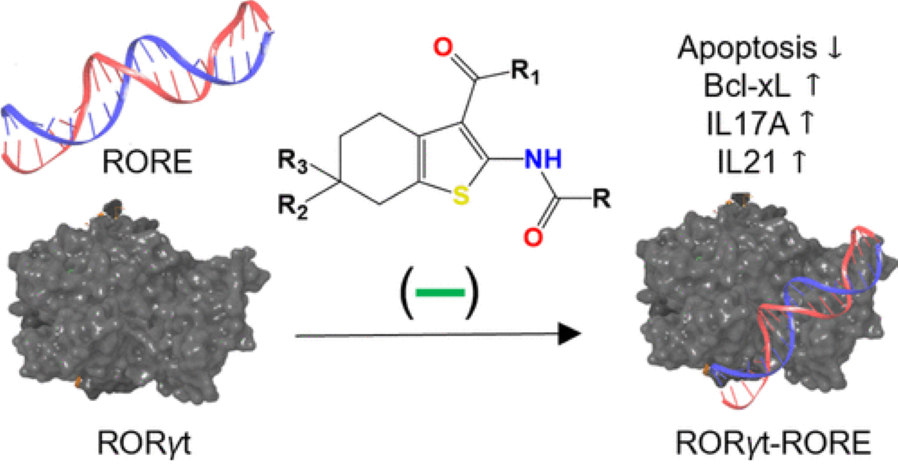
21. Discovery, Synthesis, and In Vitro Characterization of 2,3 Derivatives of 4,5,6,7-Tetrahydro-Benzothiophene as Potent Modulators of Retinoic Acid Receptor-Related Orphan Receptor γt
A. Fouda, S. Negi, O. Zaremba, R. S. Gaidar, Y. S. Moroz, E. Rusanov, S. Paraskevas, J. Tchervenkov

-
22. ZINC-22─A Free Multi-Billion-Scale Database of Tangible Compounds for Ligand Discovery
B. I. Tingle, K. G. Tang, M. Castanon, J. J. Gutierrez, M. Khurelbaatar, C. Dandarchuluun, Y. S. Moroz, J. J. Irwin

-
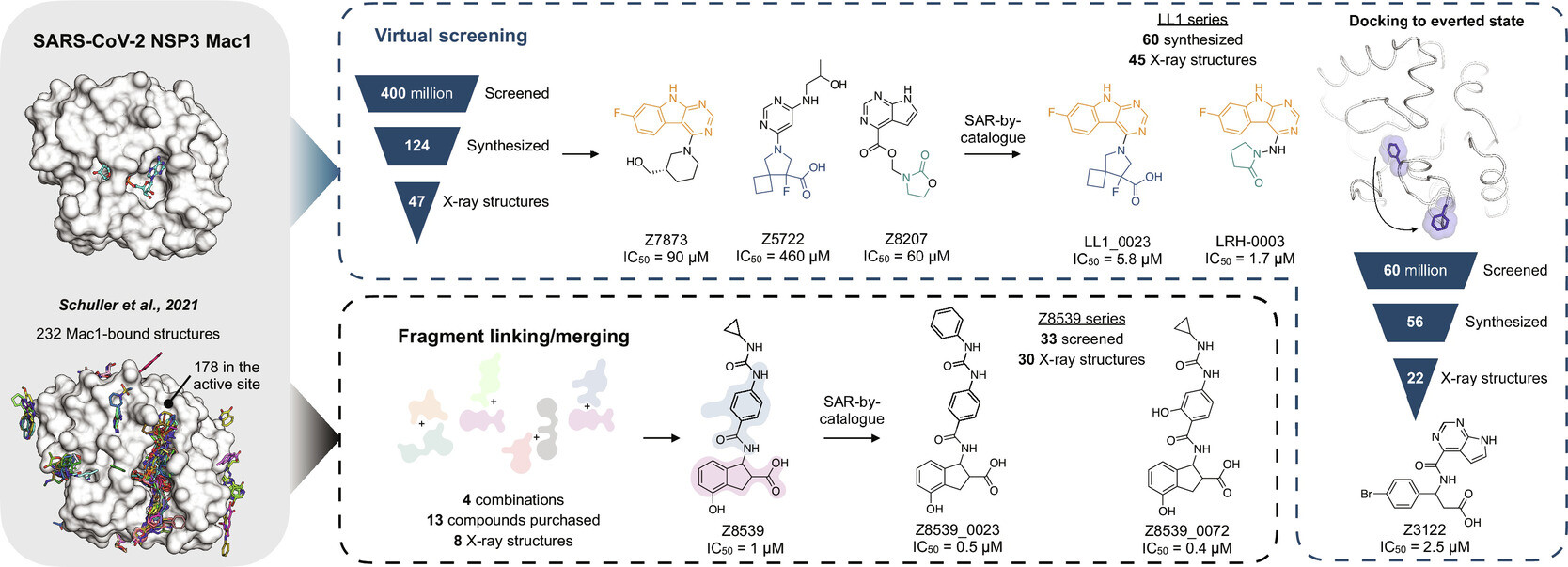
23. Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2
S. Gahbauer, G. J. Correy, M. Schuller, M. P. Ferla, Y. U. Doruk, M. Rachman, T. Wu, M. Diolaiti, S. Wang, R. J. Neitz, D. Fearon, D. S. Radchenko, Y. S. Moroz, J. J. Irwin, A. R. Renslo, J. C. Taylor, J. E. Gestwicki, F. von Delft, A. Ashworth, I. Ahel, B. K. Shoichet, J. S. Fraser

-
24. Challenges for chemistry in Ukraine after the war: Ukrainian science requires rebuilding and support
I. S. Kondratov, Y. S. Moroz, C. Gorgulla, O. O. Grygorenko, I. V. Komarov, G. Wagner, A. A. Tolmachev

-
25. C−C Coupling through Nitrogen Deletion: Application to Library Synthesis
S. Holovach, K. P. Melnykov, I. Poroshyn, R. T. Iminov, D. Dudenko, I. Kondratov, M. Levin, O. O. Grygorenko

-
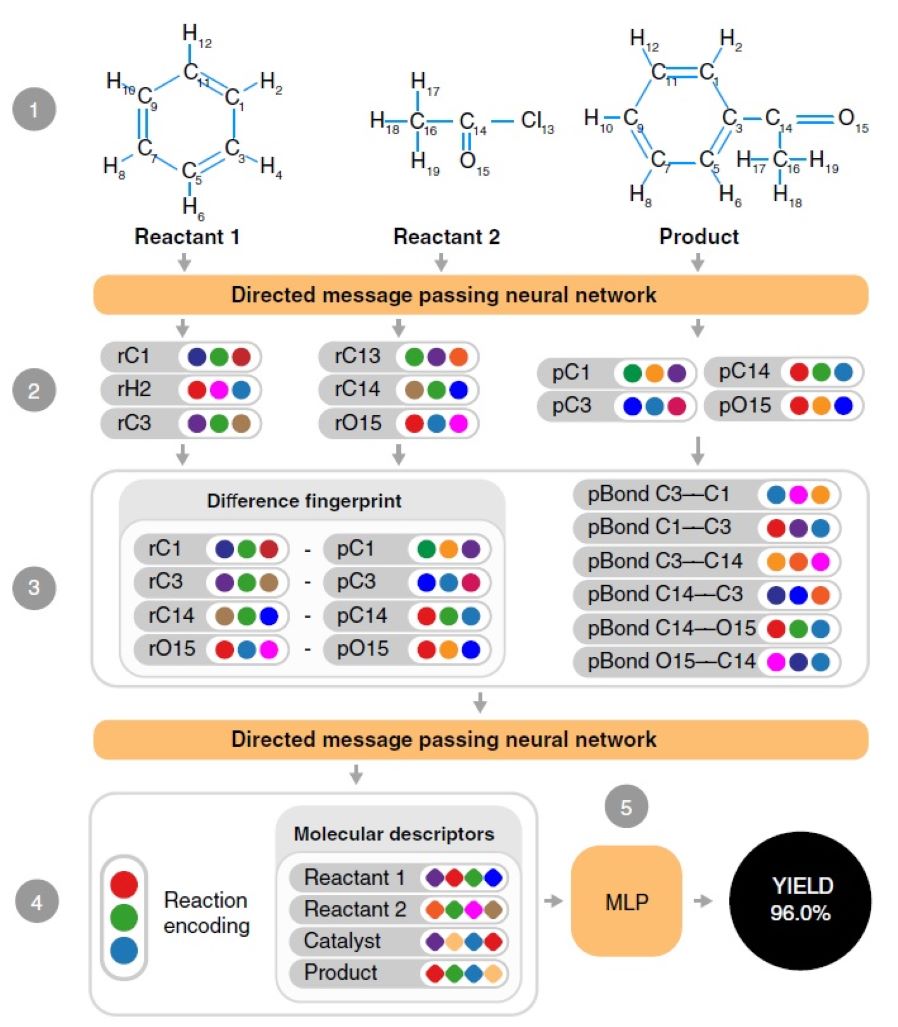
26. Advancing molecular graphs with descriptors for the prediction of chemical reaction yields
D. Yarish, S. Garkot, O. O. Grygorenko, D. S. Radchenko, Y. S. Moroz, O. Gurbych

-
27. Generative and reinforcement learning approaches for the automated de novo design of bioactive compounds
M. Korshunova, N. Huang, S. Capuzzi, D. S. Radchenko, O. Savych, Y. S. Moroz, C. I. Wells, T. M. Willson, A. Tropsha, O. Isayev
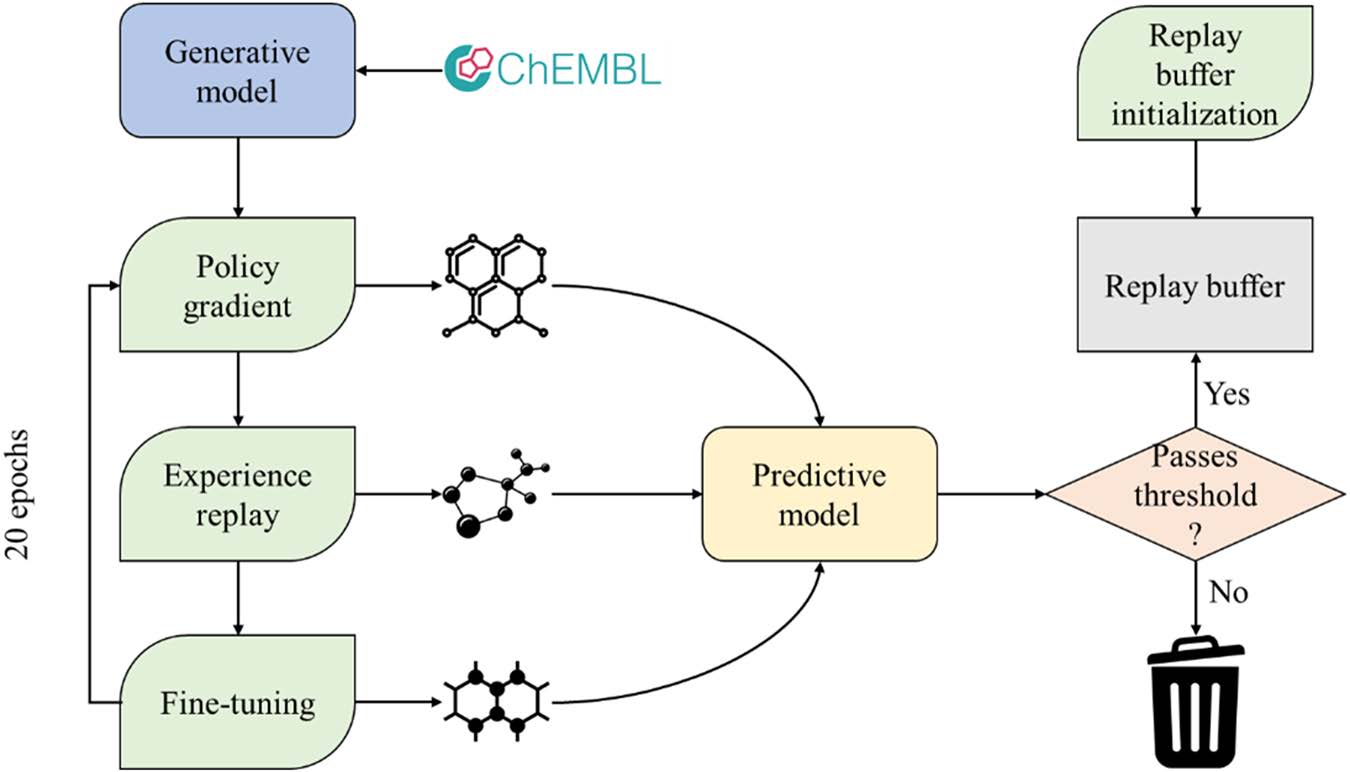

-
28. Magnet for the Needle in Haystack: “Crystal Structure First” Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces
J. Müller, R. Klein, O. Tarkhanova, A. Gryniukova, P. Borysko, S. Merkl, M. Ruf, A. Neumann, M. Gastreich, Y. S. Moroz, G. Klebe, S. Glinca
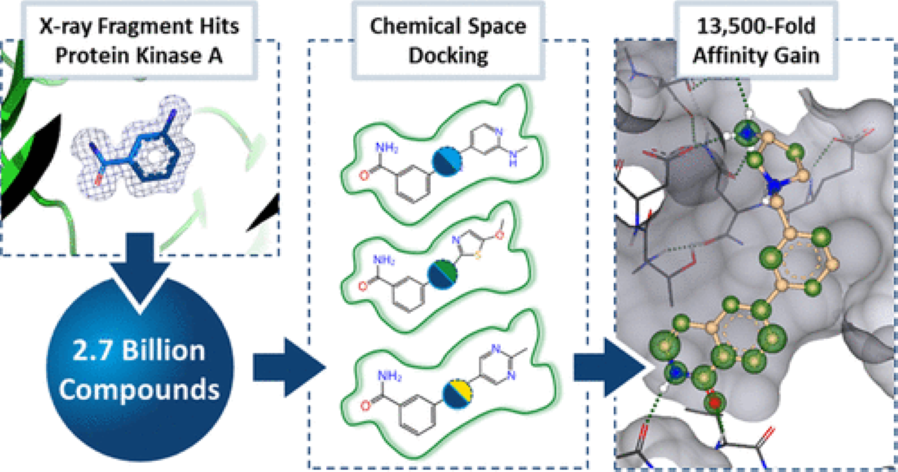

-
29. Structure-based discovery of nonopioid analgesics acting through the α2A-adrenergic receptor
E. A. Fink, J. Xu, H. Hübner, J. M. Braz, P. Seemann, C. Avet, V. Craik, D. Weikert, M. F. Schmidt, C. M. Webb, N. A. Tolmachova, Y. S. Moroz, X.-P. Huang, C. Kalyanaraman, S. Gahbauer, G. Chen, Z. Liu, M. P. Jacobson, J. J. Irwin, M. Bouvier, Y. Du, B. K. Shoichet, A. I. Basbaum, P. Gmeiner
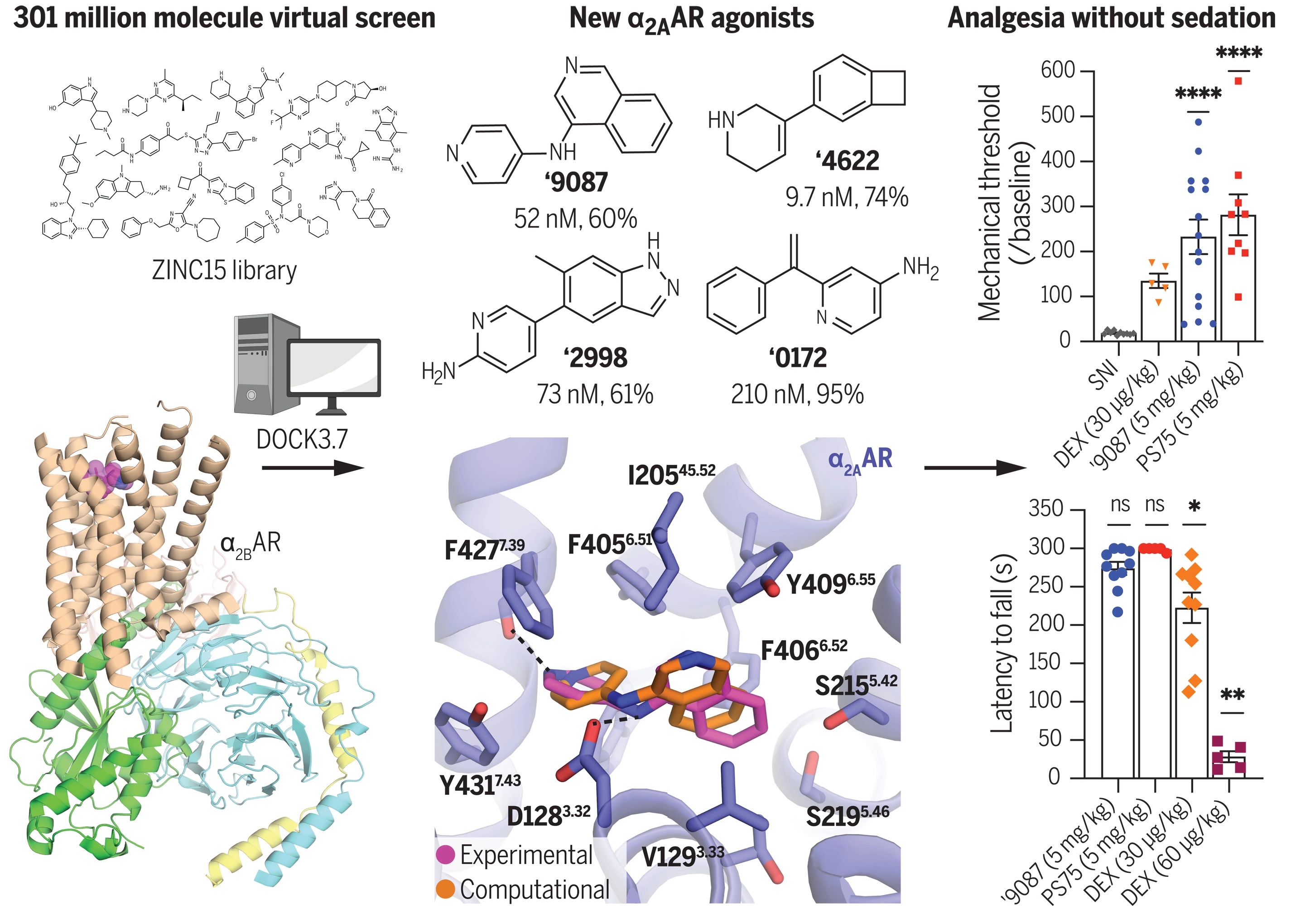

-
30. The Ukrainian Factor in Early-Stage Drug Discovery in the Context of Russian Invasion: The Case of Enamine Ltd
I. S. Kondratov, Y. S. Moroz, O. O. Grygorenko, A. A. Tolmachev


-
31. In Vitro and In Silico Evaluation of Cholinesterase Inhibition by Alkaloids Obtained from Branches of Abuta panurensis Eichler
R. da Silva Mesquita, A. Kyrylchuk, A. Cherednichenko, I. S. Costa Sá, L. Macedo Bastos, F. Moura Araújo da Silva, R. de C. Saraiva Nunomura, A. Grafov


-
32. Creation of targeted compound libraries based on 3D shape recognition
A. Kyrylchuk, I. Kravets, A. Cherednichenko, V. Tararina, A. Kapeliukha, D. Dudenko, M. Protopopov


-
33. Virtual Screening in Search for a Chemical Probe for Angiotensin-Converting Enzyme 2 (ACE2)
I. O. Kravets, D. V. Dudenko, A. E. Pashenko, T. A. Borisova, G. M. Tolstanova, S. V. Ryabukhin, D. M. Volochnyuk


-
34. A Close-up Look at the Chemical Space of Commercially Available Building Blocks for Medicinal Chemistry
Y. Zabolotna, D. M. Volochnyuk, S. V. Ryabukhin, D. Horvath, K. S. Gavrilenko, G. Marcou, Y. S. Moroz, O. Oksiuta, A. Varnek


-
35. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds
A. A. Sadybekov, A. V. Sadybekov, Y. Liu, C. Iliopoulos-Tsoutsouvas, X.-P. Huang, J. Pickett, B. Houser, N. Patel, N. K. Tran, F. Tong, N. Zvonok, M. K. Jain, O. Savych, D. S. Radchenko, S. P. Nikas, N. A. Petasis, Y. S. Moroz, B. L. Roth, A. Makriyannis, V. Katritch


-
36. Structures of the σ2 receptor enable docking for bioactive ligand discovery
A. Alon, J. Lyu, J. M. Braz, T. A. Tummino, V. Craik, M. J. O’Meara, C. M. Webb, D. S. Radchenko, Y. S. Moroz, X.-P. Huang, Y. Liu, B. L. Roth, J. J. Irwin, A. I. Basbaum, B. K. Shoichet, A. C. Kruse


-
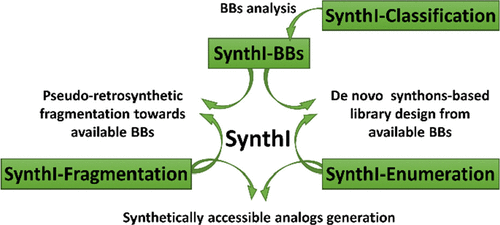
37. SynthI: A New Open-Source Tool for Synthon-Based Library Design
Y. Zabolotna, D. M. Volochnyuk, S. V. Ryabukhin, K. Gavrylenko, D. Horvath, O. Klimchuk, O. Oksiuta, G. Marcou, A. Varnek
-
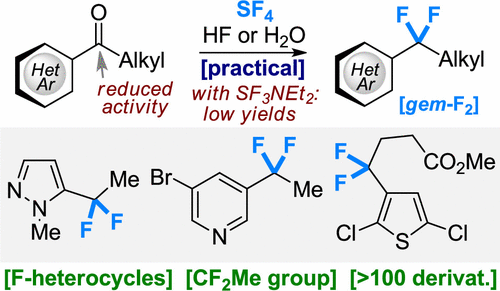
38. Scalable Approach to Fluorinated Heterocycles with Sulfur Tetrafluoride (SF4)
S. Trofymchuk, M. Bugera, A. A. Klipkov, V. Ahunovych, B. Razhyk, S. Semenov, A. Boretskyi, K. Tarasenko, P. K. Mykhailiuk
-
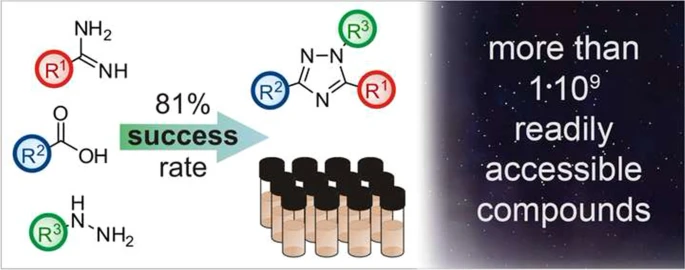
39. One-pot parallel synthesis of 1,3,5-trisubstituted 1,2,4-triazoles
D. S. Radchenko, V. S. Naumchyk, I. Dziuba, A. A. Kyrylchuk, K. E. Gubina, Y. S. Moroz, O. O. Grygorenko
-
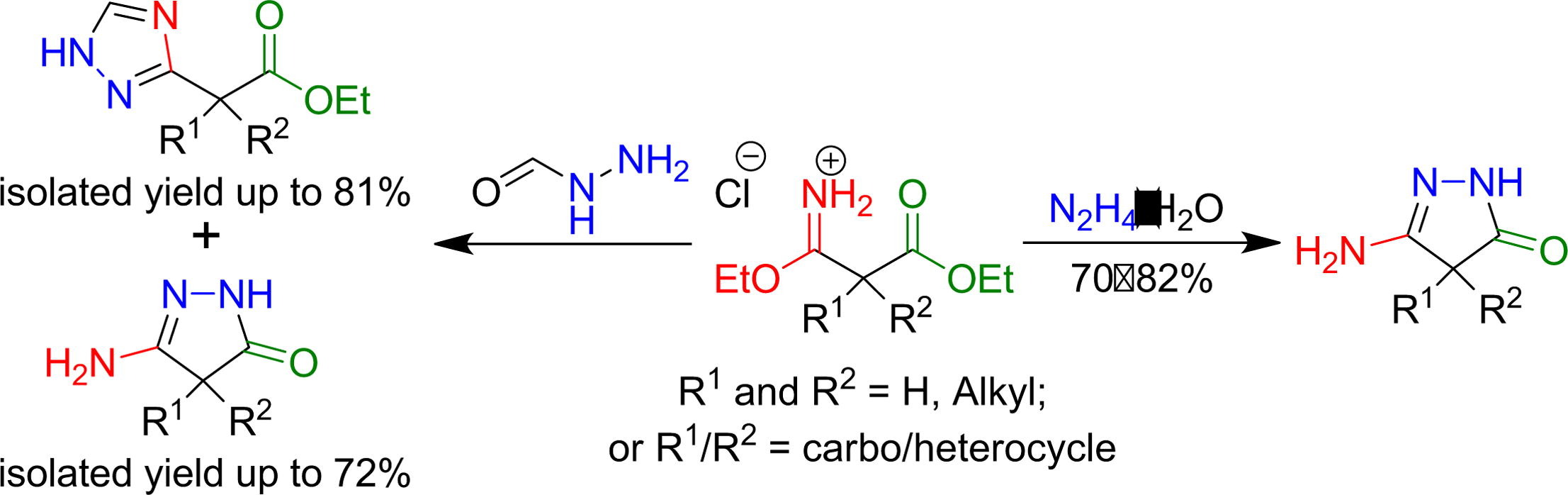
40. Synthesis of α-substituted 2-(1H-1,2,4-triazol-3-yl)acetates and 5-amino-2,4-dihydro-3H-pyrazol-3-ones via the Pinner strategy
D. M. Khomenko, R. O. Doroshchuk, H. V. Ivanova, B. V. Zakharchenko, I. V. Raspertova, O. V. Vaschenko, S. Shova, A. V. Dobrydnev, Y. S. Moroz, O. O. Grygorenko, R. D. Lampeka
-
41. A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening
C. Gorgulla, K. M. Padmanabha Das, K. E. Leigh, M. Cespugli, P. D. Fischer, Z.-F. Wang, G. Tesseyre, S. Pandita, A. Shnapir, A. Calderaio, M. Gechev, A. Rose, N. Lewis, C. Hutcheson, E. Yaffe, R. Luxenburg, H. D. Herce, V. Durmaz, T. D. Halazonetis, K. Fackeldey, J. J. Patten, A. Chuprina, I. Dziuba, A. Plekhova, Y. Moroz, D. Radchenko, O. Tarkhanova, I. Yavnyuk, C. Gruber, R. Yust, D. Payne, A. M. Näär, M. N. Namchuk, R. A. Davey, G. Wagner, J. Kinney, H. Arthanari
-
42. Generating Multibillion Chemical Space of Readily Accessible Screening Compounds
O. O. Grygorenko, D. S. Radchenko, I. Dziuba, A. Chuprina, K. E. Gubina, Y. S. Moroz
-
43. SAVI, in silico generation of billions of easily synthesizable compounds through expert-system type rules
H. Patel, W.-D. Ihlenfeldt, P. N. Judson, Y. S. Moroz, Y. Pevzner, M. L. Peach, V. Delannée, N. I. Tarasova, M. C. Nicklaus
-
44. ZINC20—A Free Ultralarge-Scale Chemical Database for Ligand Discovery
J. J. Irwin, K. G. Tang, J. Young, C. Dandarchuluun, B. R. Wong, M. Khurelbaatar, Y. S. Moroz, J. Mayfield, R. A. Sayle
-
45. Atomic ring invariant and Modified CANON extended connectivity algorithm for symmetry perception in molecular graphs and rigorous canonicalization of SMILES
D. G. Krotko
-
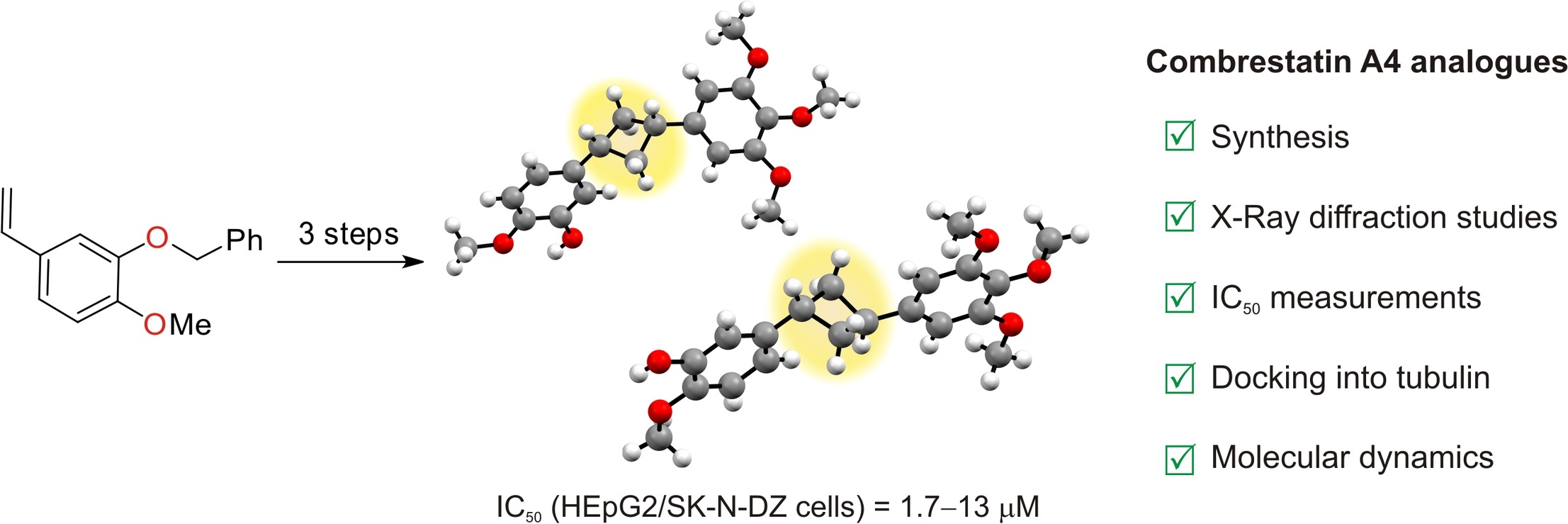
46. Synthesis, biological evaluation, and modeling studies of 1,3-disubstituted cyclobutane-containing analogs of combretastatin A4
A. Malashchuk, A. V. Chernykh, V. V. Hurmach, M. O. Platonov, O. Onopchenko, S. Zozulya, C. G. Daniliuc, A. V. Dobrydnev, I. S. Kondratov, Y. S. Moroz, O. O. Grygorenko
-
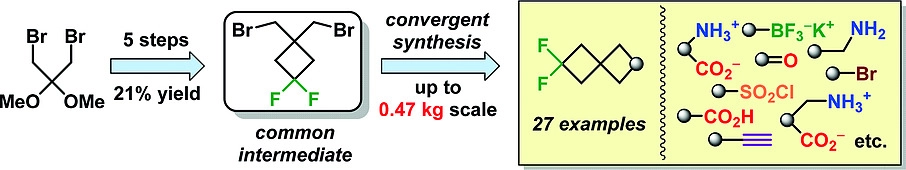
47. Multigram Synthesis of Advanced 6,6-Difluorospiro[3.3]heptane-Derived Building Blocks
S. Olifir, A. V. Chernykh, A. V. Dobrydnev, O. O. Grygorenko, Y. S. Moroz, Z. V. Voitenko, D. S. Radchenko
-
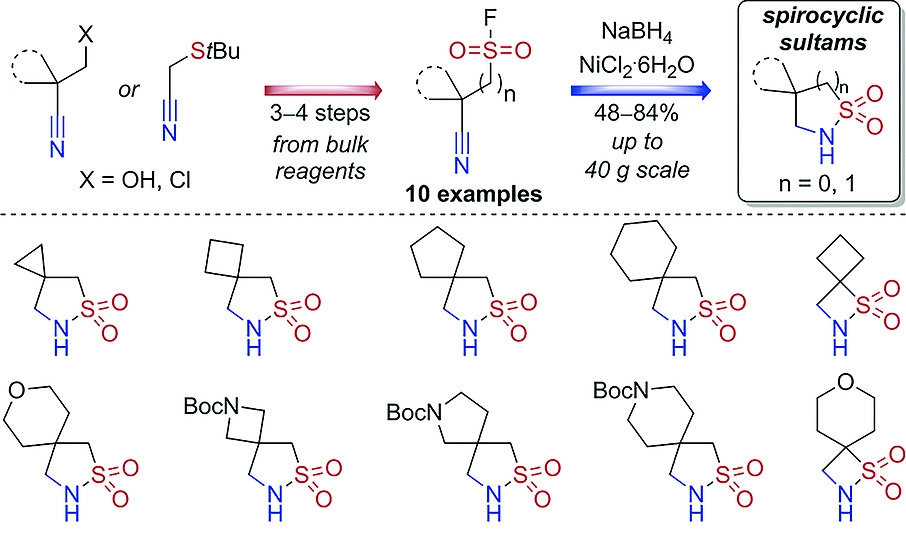
48. Synthesis of Spirocyclic β‐ and γ‐Sultams by One‐Pot Reductive Cyclization of Cyanoalkylsulfonyl Fluorides
K. O. Stepannikova, B. V. Vashchenko, O. O. Grygorenko, M. V. Gorichko, A. Y. Cherepakha, Y. S. Moroz, Y. M. Volovenko, S. Zhersh
-
49. An open-source drug discovery platform enables ultra-large virtual screens
C. Gorgulla, A. Boeszoermenyi, Z.-F. Wang, P. D. Fischer, P. W. Coote, K. M. Padmanabha Das, Y. S. Malets, D. S. Radchenko, Y. S. Moroz, D. A. Scott, K. Fackeldey, M. Hoffmann, I. Iavniuk, G. Wagner, H. Arthanari
-
50. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms
R. M. Stein, H. J. Kang, J. D. McCorvy, G. C. Glatfelter, A. J. Jones, T. Che, S. Slocum, X.-P. Huang, O. Savych, Y. S. Moroz, B. Stauch, L. C. Johansson, V. Cherezov, T. Kenakin, J. J. Irwin, B. K. Shoichet, B. L. Roth, M. L. Dubocovich
-
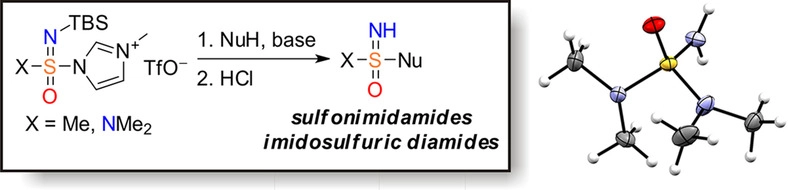
51. Sulfonimidamides and Imidosulfuric Diamides: Compounds from an Underexplored Part of Biologically Relevant Chemical Space
S. V. Zasukha, V. M. Timoshenko, A. A. Tolmachev, V. O. Pivnytska, O. Gavrylenko, S. Zhersh, Y. Shermolovich, O. O. Grygorenko
-
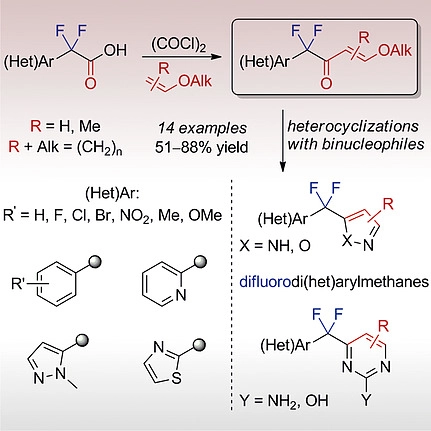
52. (Het)aryl Difluoromethyl-Substituted β-Alkoxyenones: Synthesis and Heterocyclizations
Y. Bugera, K. V. Tarasenko, I. S. Kondratov, I. I. Gerus, B. V. Vashchenko, V. E. Ivasyshyn, O. O. Grygorenko
-
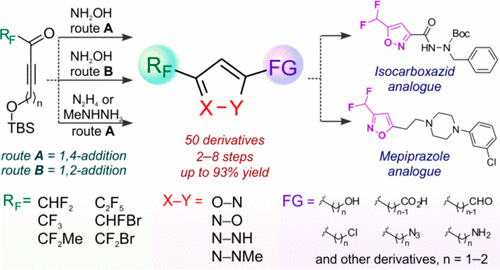
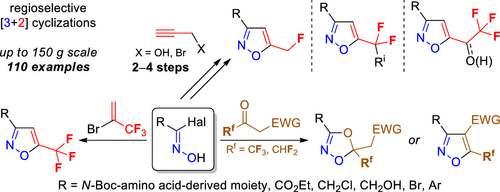
53. Regioselective Synthesis of Functionalized 3- or 5-Fluoroalkyl Isoxazoles and Pyrazoles from Fluoroalkyl Ynones and Binucleophiles
B. A. Chalyk, A. Khutorianskyi, A. Lysenko, Y. Fil, Y. O. Kuchkovska, K. S. Gavrilenko, I. Bakanovych, Y. S. Moroz, A. O. Gorlova, O. O. Grygorenko
-
54. Synthesis of 5-(Fluoroalkyl)isoxazole Building Blocks by Regioselective Reactions of Functionalized Halogenoximes
B. A. Chalyk, K. V. Hrebeniuk, Y. V. Fil, K. S. Gavrilenko, A. B. Rozhenko, B. V. Vashchenko, O. V. Borysov, A. V. Biitseva, P. S. Lebed, I. Bakanovych, Y. S. Moroz, O. O. Grygorenko
-
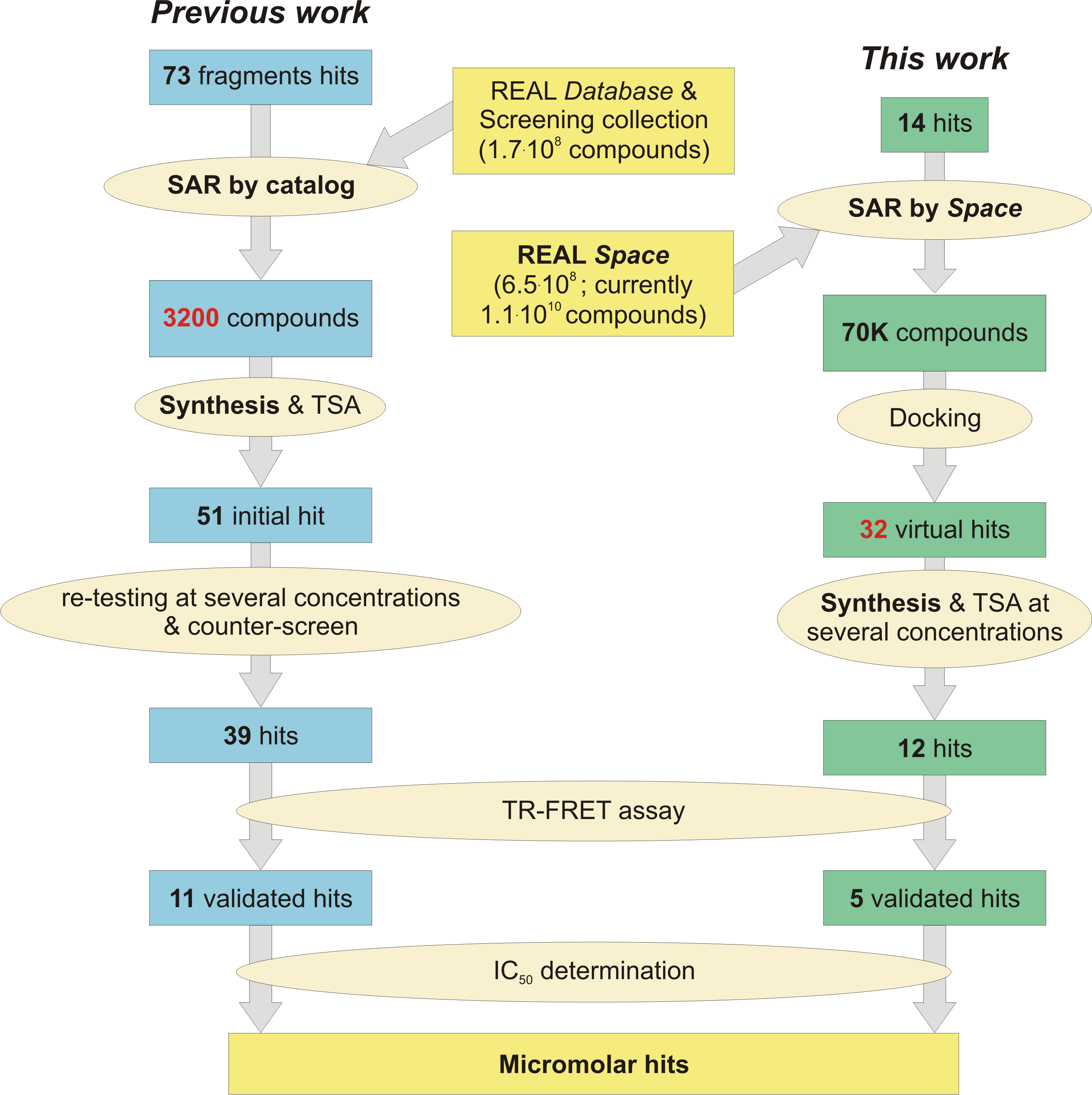
55. SAR by Space: Enriching Hit Sets from the Chemical Space
F.-M. Klingler, M. Gastreich, O. Grygorenko, O. Savych, P. Borysko, A. Griniukova, K. Gubina, C. Lemmen, Y. Moroz
-
56. One-Pot Parallel Synthesis of 5-(Dialkylamino)tetrazoles
O. Savych, Y. O. Kuchkovska, A. V. Bogolyubsky, A. I. Konovets, K. E. Gubina, S. E. Pipko, A. V. Zhemera, A. V. Grishchenko, D. N. Khomenko, V. S. Brovarets, R. Doroschuk, Y. S. Moroz, O. O. Grygorenko
-
57. Pros and cons of virtual screening based on public ‘Big Data’: In silico mining for new bromodomain inhibitors
I. Casciuc, D. Horvath, A. Gryniukova, K. A. Tolmachova, O. V. Vasylchenko, P. Borysko, Y. S. Moroz, J. Bajorath, A. Varnek
-
58. Ultra-large library docking for discovering new chemotypes
J. Lyu, S. Wang, T. E. Balius, I. Singh, A. Levit, Y. S. Moroz, M. J. O’Meara, T. Che, E. Algaa, K. Tolmachova, A. A. Tolmachev, B. K. Shoichet, B. L. Roth, J. J. Irwin
-
59. Evolution of commercially available compounds for HTS
D. M. Volochnyuk, S. V. Ryabukhin, Y. S. Moroz, O. Savych, A. Chuprina, D. Horvath, Y. Zabolotna, A. Varnek, D. B. Judd
-
60. (Chlorosulfonyl)benzenesulfonyl Fluorides—Versatile Building Blocks for Combinatorial Chemistry: Design, Synthesis and Evaluation of a Covalent Inhibitor Library
K. A. Tolmachova, Y. S. Moroz, A. Konovets, M. O. Platonov, O. V. Vasylchenko, P. Borysko, S. Zozulya, A. Gryniukova, A. V. Bogolubsky, S. Pipko, P. K. Mykhailiuk, V. S. Brovarets, O. O. Grygorenko
-
61. Facile One-Pot Parallel Synthesis of 3-Amino-1,2,4-triazoles
A. V. Bogolyubsky, O. Savych, A. V. Zhemera, S. E. Pipko, A. V. Grishchenko, A. I. Konovets, R. O. Doroshchuk, D. N. Khomenko, V. S. Brovarets, Y. S. Moroz, M. Vybornyi
-
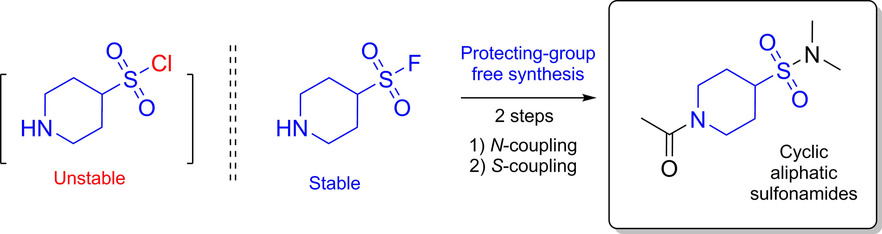
62. Saturated Heterocyclic Aminosulfonyl Fluorides: New Scaffolds for Protecting-Group-Free Synthesis of Sulfonamides
S. A. Zhersh, O. P. Blahun, I. V. Sadkova, A. A. Tolmachev, Y. S. Moroz, P. K. Mykhailiuk
-
1. A Benchmark Set of Bioactive Molecules for Diversity Analysis of Compound Libraries and Combinatorial Chemical Spaces
Alexander Neumann, Raphael Klein
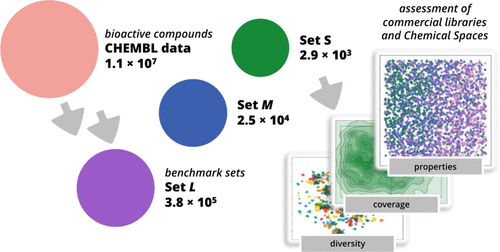

-
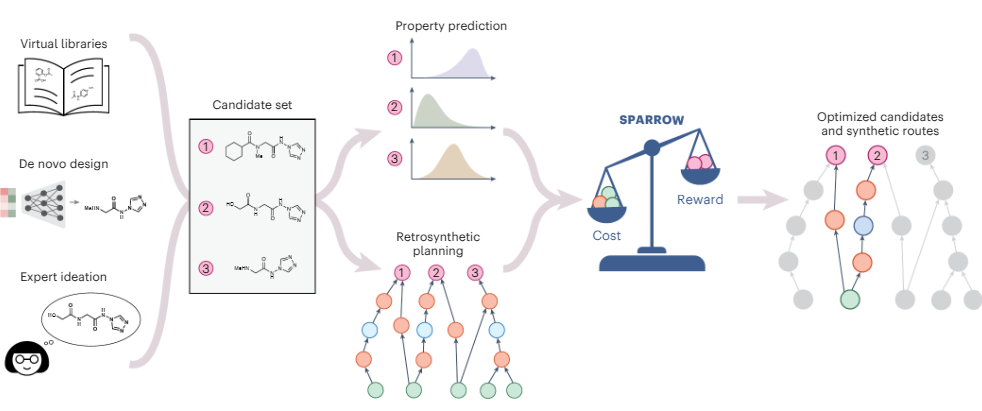
2. An algorithmic framework for synthetic cost-aware decision making in molecular design
Jenna C. Fromer, Connor W. Coley

-
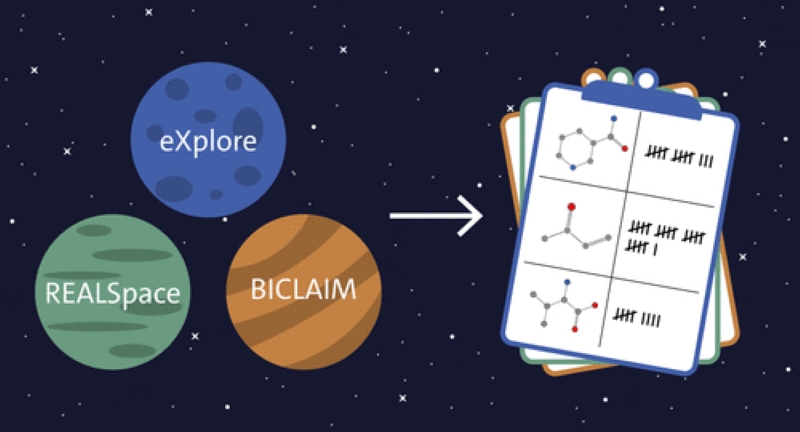
3. Enhanced Calculation of Property Distributions in Chemical Fragment Spaces
Justin Lübbers, Uta Lessel, and Matthias Rarey

-
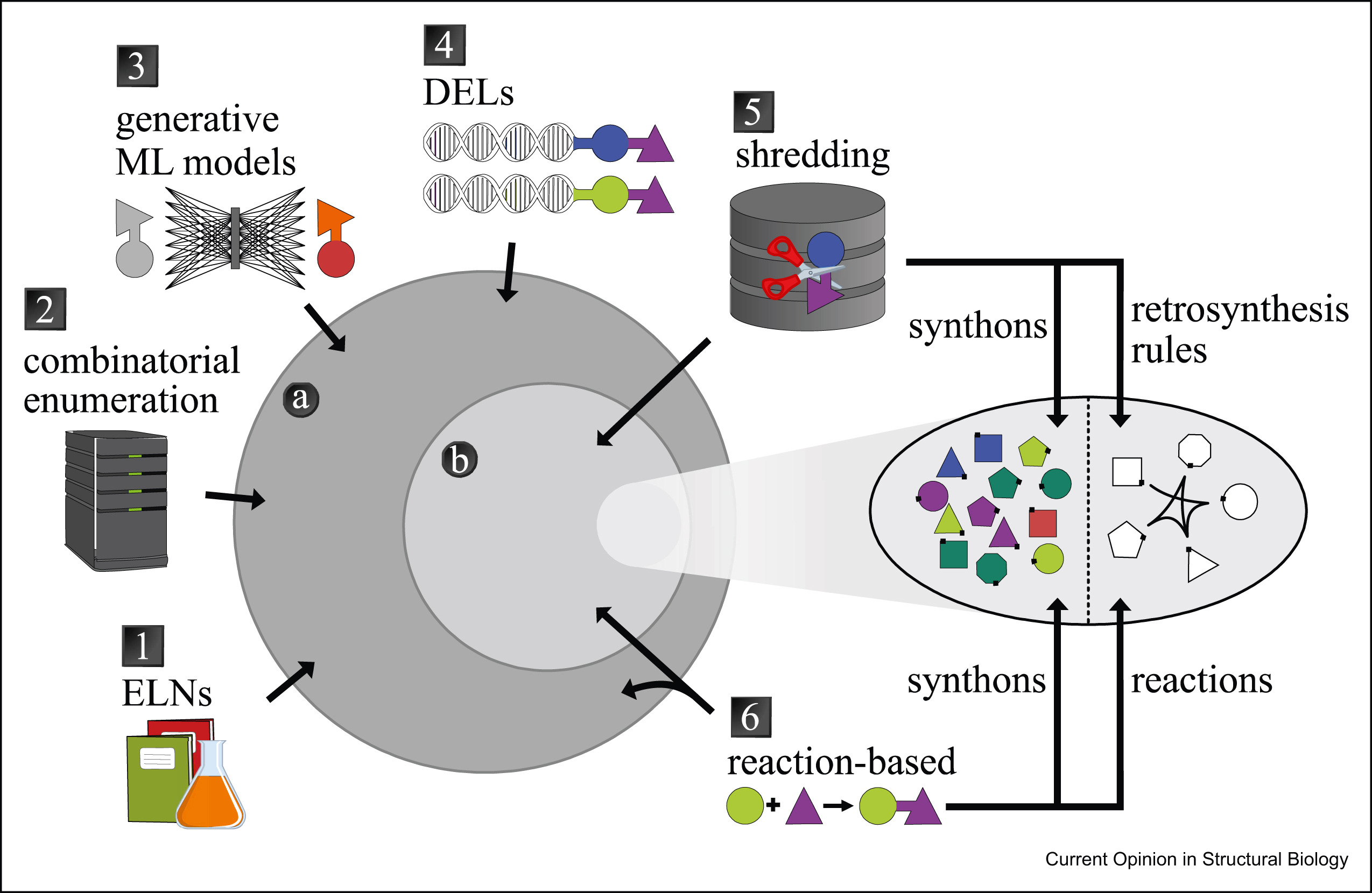
4. Navigating large chemical spaces in early-phase drug discovery
M. Korn, C. Ehrt, F. Ruggiu, M. Gastreich, M. Rarey

-
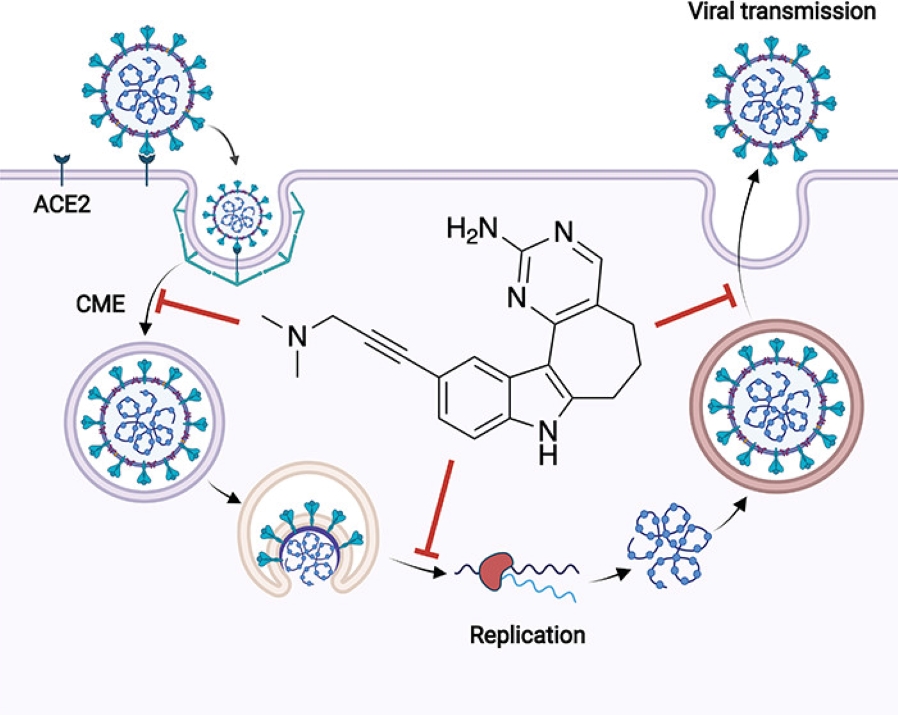
5. Identification and Utilization of a Chemical Probe to Interrogate the Roles of PIKfyve in the Lifecycle of β-Coronaviruses
D. H. Drewry, F. M. Potjewyd, A. Bayati, J. L. Smith, R. J. Dickmander, S. Howell, S. Taft-Benz, S. M. Min, M. A. Hossain, M. Heise, P. S. McPherson, N. J. Moorman, A. D. Axtman

-
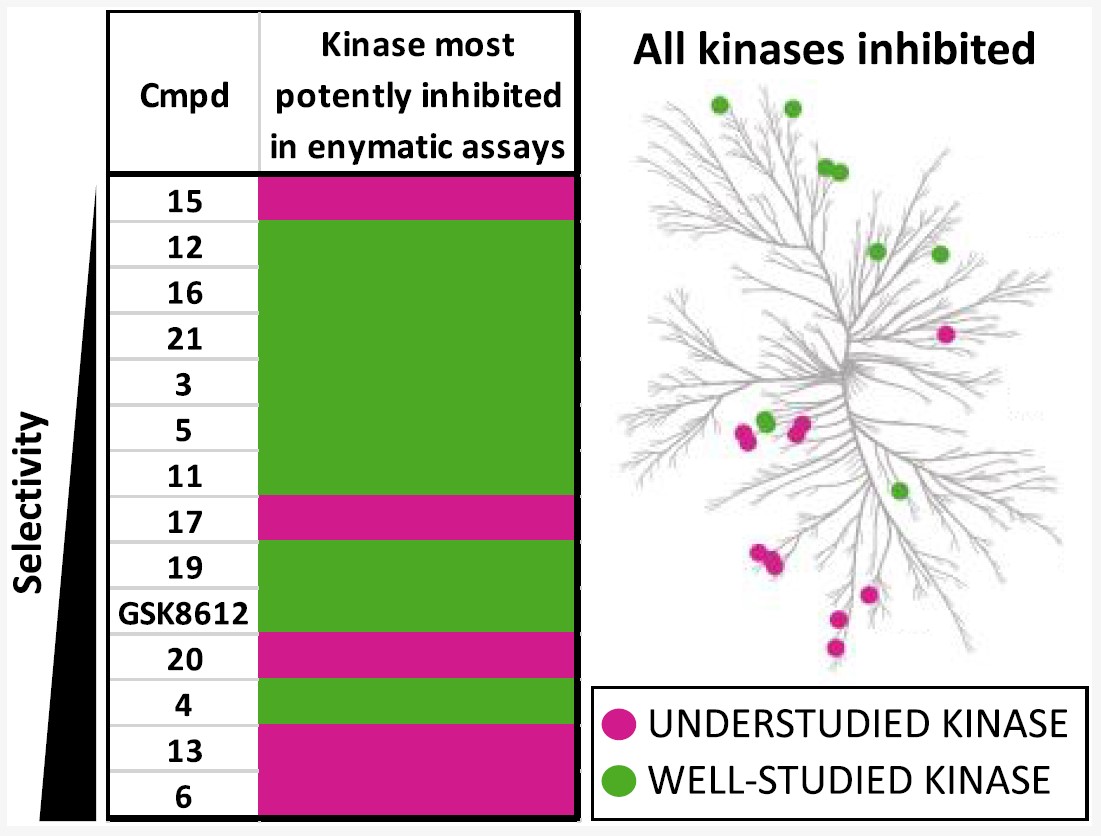
6. Identification of Pyrimidine-Based Lead Compounds for Understudied Kinases Implicated in Driving Neurodegeneration
D. H. Drewry, J. K. Annor-Gyamfi, C. I. Wells, J. E. Pickett, V. Dederer, F. Preuss, S. Mathea, A. D. Axtman
-
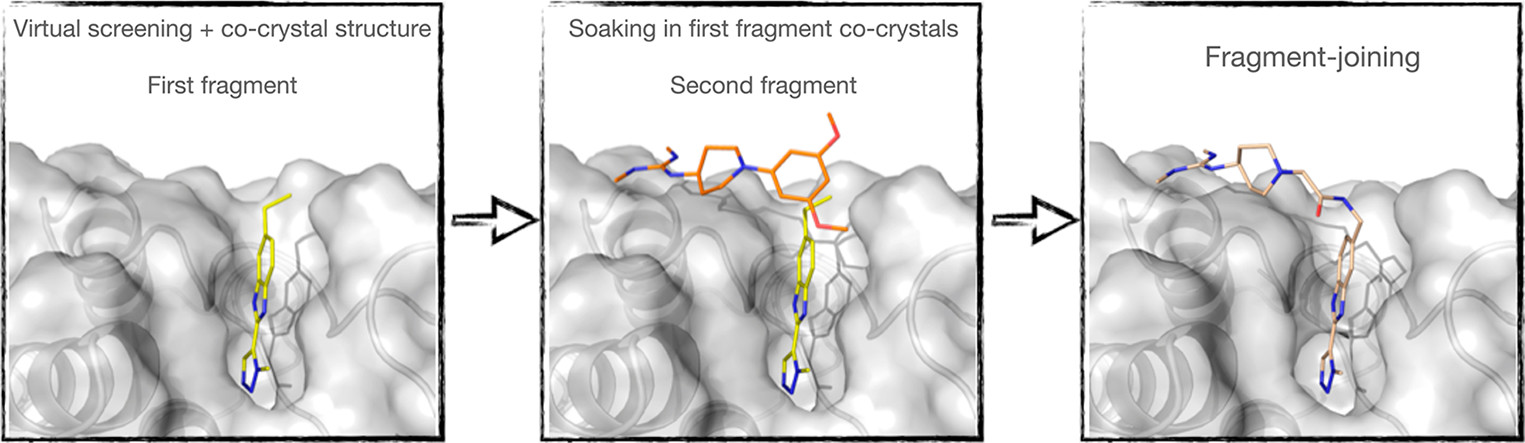
7. Identification of a BAZ2A Bromodomain Hit Compound by Fragment Joining
A. Dalle Vedove, G. Cazzanelli, J. Corsi, M. Sedykh, V. G. D’Agostino, A. Caflisch, G. Lolli
-
8. Degradation Characteristics of a Novel PAF Receptor Antagonist, SY0916, in Aqueous Solution
B. Jin, Y. Wang, T. Zhang, W. Yin, D. Zhang, H. Huang, C. Ma