About Protein Kinases
Since the 1980s, protein kinases have been recognized as potential drug targets, particularly based on advances in the molecular understanding of cancer.
Approximately 110 novel kinases are currently being explored as targets, which together with 45 targets of approved kinase inhibitors represent only about 30% of the human kinome, indicating that there are still substantial unexplored opportunities for this drug class.*
General description of process
Chemspace computational services team prepared structurally diverse Protein Kinases Targeted Library based on USRCAT similarity.** For a better understanding of the approach, we created a description available for downloading. It gives brief information on Protein Kinases, USRCAT, the algorithm and its validation, explains the process of creating the library.
Some words about USRCAT and how it works
USRCAT (The Ultrafast Shape Recognition with CREDO Atom Types) is a Ligand-Based Virtual Screening (LBVS) method.** This method is capable of retrieving compounds with sharing 3D molecular shape with minimal topological similarity. This means the possibility of finding compounds, that will be completely different by structure but have high potential interaction with the corresponding receptor.
USRCAT condenses 3-dimensional information about molecular shape, as well as other properties, into a small set of numeric descriptors. This gives the ability to compare molecules fast and accurately.
What we offer
As a result of applying steps indicated in the workflow to a set of reference compounds for different targets, we prepared two sets of screening compounds from the Chemspace catalog:
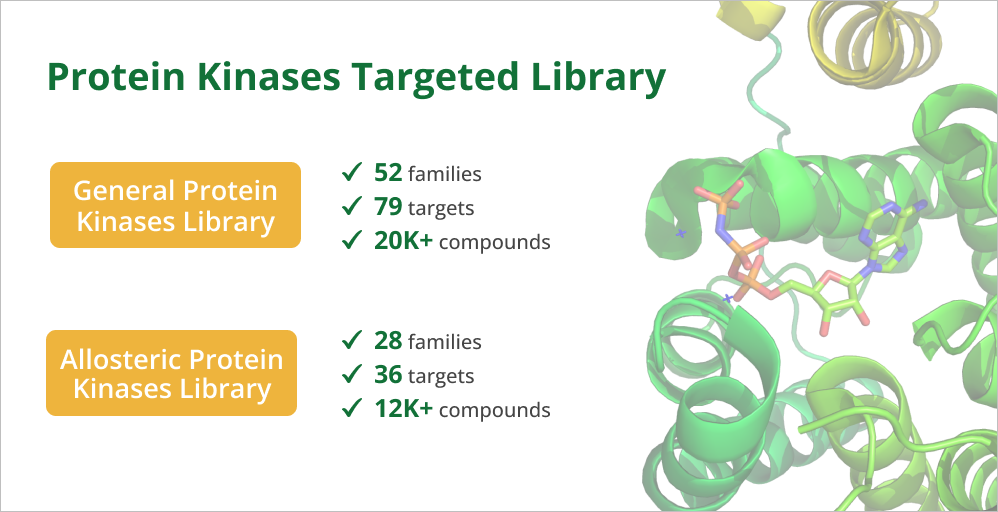
1. General Protein Kinases Library - prepared based on the list of targets and ligands from the publication.* It contains 20K+ compounds against 79 targets belonging to 52 families.
2. Allosteric Protein Kinases Library - prepared based on the information from KLIFS.net - kinase database.*** It contains 12K+ compounds against 36 targets belonging to 28 families.
You can find more information on the targets in the attached Excel file, that we prepared for you.
Custom libraries design
If you did not find the desired target, please feel free to request library generation.
You can find the full list of targets in the attachment on the Protein Kinases Targeted Libraries.
We offer generation targeted libraries using known ligands and analog similarity search by USRCAT (method to search for new chemotypes), as well as methods with Morgan FP and E3FP. Besides that, docking services are available upon request.
To find out more or request a custom library drop an e-mail at: [email protected]
* Attwood, M. M., Fabbro, D., Sokolov, A. V., Knapp, S., & Schiöth, H. B. (2021). Trends in kinase drug discovery: targets, indications and inhibitor design. Nature Reviews Drug Discovery. doi:10.1038/s41573-021-00252-y
**Schreyer, A. M., & Blundell, T. (2012). USRCAT: real-time ultrafast shape recognition with pharmacophoric constraints. Journal of cheminformatics, 4(1), 27. doi:10.1186/1758-2946-4-27
*** Georgi K Kanev, Chris de Graaf, Bart A Westerman, Iwan J P de Esch, Albert J Kooistra, KLIFS: an overhaul after the first 5 years of supporting kinase research, Nucleic Acids Research, Volume 49, Issue D1, 8 January 2021, Pages D562–D569, doi:10.1093/nar/gkaa895