V-SYNTHES Approach
Enamine xREAL Space currently contains 4.4T molecules and has the potential to expand even further, which calls for new virtual screening methods that can be used with chemical spaces of such size. This problem is now covered by V-SYNTHES MolSoft ICM-Pro, the virtual synthon hierarchical enumeration screening approach, that allows screening of the chemical spaces of billions and trillions of compounds without full enumeration [1].
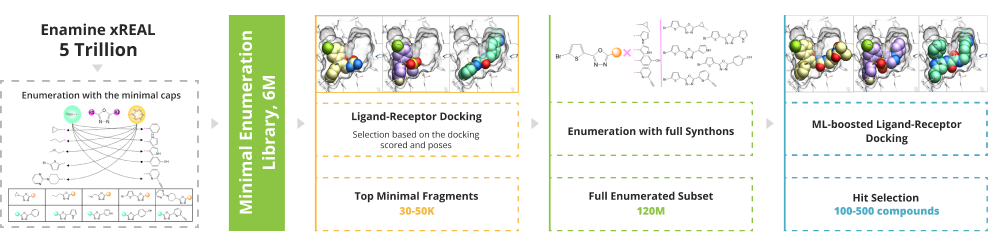
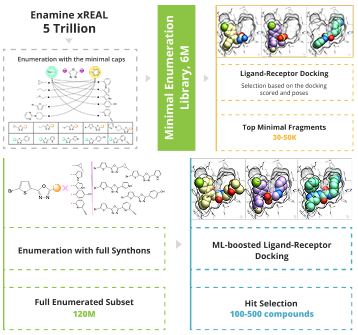
To enable the giga-scale virtual screening the minimal enumeration library (MEL) should be designed. MEL consists of the Enamine REAL synthons capped with minimal counter synthon and represents the chemical diversity of the whole space. MEL is used to identify the capped synthons that anchored the pocket. These synthons are enumerated utilizing the well-validated Enamine REAL reactions to generate the focused synthetically accessible space. The molecular docking is done using the state-of-the-art software ICM-Pro by MolSoft .
Example of Enumeration in 3D
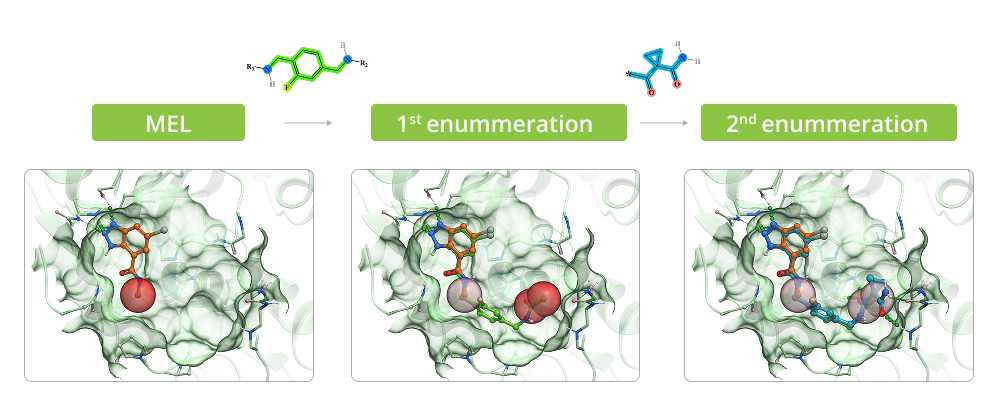
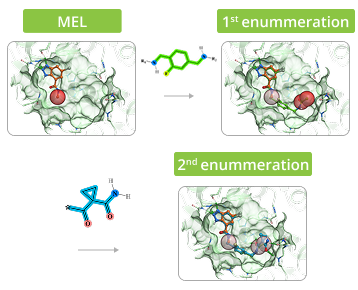
The process of enumeration follows molecular docking of minimum enumerated library (MEL) into target protein and identification of top-scored fragments based on acquired data. MEL is a library of fragment-like compounds that represent all possible scaffold-synthon combinations for all the reactions in the whole Enamine REAL Space. The library is built from the reaction scaffolds, enumerated with corresponding synthons at one of its R positions whilst all other R positions are capped with minimal synthon (usually Me or Phe group) according to the reaction type for a given R position. Enumeration step involves iterative replacement of capped R group with a full range of corresponding synthons from the library. Number of iterations depends on reaction type: for two-component reactions it is a single-step process, whereas for three-component reactions it requires two steps. Full-enumerated compounds are then docked into the same protein pocket to prove their ability to fit in there in the same binding mode that was observed for initial fragment extracted from MEL.
Project Requirements
For us to complete this workflow on your target of interest, a well-defined protein structure must be available (X-ray, cryo-EM, NMR, homology model).
Additional requirements for hit selection can be discussed upon request.
Workflow Description
The project starts with the development of a docking model based on your protein of interest. We can help with the target analysis by evaluating the quality of the protein structure, selecting the pocket to target, and building the ensemble of pocket confirmations to proceed with docking.
The first step of V-SYNTHES MolSoft ICM-Pro is MEL docking. The high-scoring fragments will undergo CapSelect filtering to select the fragments that can be grown within the pocket. Visual inspection and occupancy analysis will close the stage of selecting the fragments for further enumeration.
Next, a new space is enumerated based on the high-scoring fragments, 1-2M of diverse compounds from this space are docked to the target of interest with higher effort.
Hit selection step is performed through docking the focused space followed by re-docking of top-ranked molecules with a higher docking effort, energy minimization for each ligand-protein complex, and utilizing AI-based RTCNN scoring function. Distance analysis towards the key residues, pocket occupancy analysis, and synthon-based clustering are used to select the final molecules for testing.
After the final hit selection step, you will be provided with the list of 100-500 compounds recommended for wet screening.
- Sadybekov, A. A.; Sadybekov, A. V.; Liu, Y.; Iliopoulos-Tsoutsouvas, C.; Huang, X.-P.; Pickett, J.; Houser, B.; Patel, N.; Tran, N. K.; Tong, F.; Zvonok, N.; Jain, M. K.; Savych, O.; Radchenko, D. S.; Nikas, S. P.; Petasis, N. A.; Moroz, Y. S.; Roth, B. L.; Makriyannis, A.; Katritch, V. Synthon-based Ligand Discovery in Virtual Libraries of Over 11 Billion Compounds. Nature 2022, 601, 452–459. https://www.nature.com/articles/s41586-021-04220-9 .