Virtual Screening
Virtual screening (VS) is a computational approach integral to drug discovery, serving as
a cost-effective and efficient method for identifying potential hits. By simulating
molecular interactions in silico, virtual screening allows researchers to prioritize and
filter through numerous compounds, significantly reducing the number of candidates that need
to be experimentally tested [1, 2, 3].
There are two main approaches inside VS: structure-based VS (SBVS) and ligand-based VS (LBVS) [3].
At Chemspace we offer numerous services for both approaches to accelerate your Drug Discovery projects.
Structure-Based Virtual Screening (SBVS)
SBVS or target-based VS (TBVS) attempts to predict the binding orientation of ligand (organic small molecule) and target molecule (protein or other biomacromolecule). The main requirement for this technique is presenting of good enough 3D structure of the target molecule. For SBVS we offer: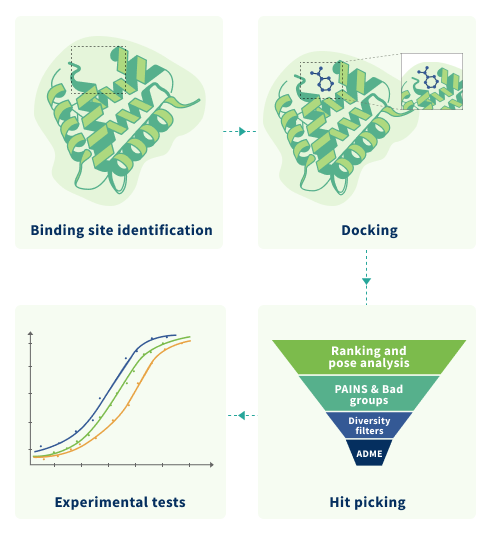
Molecular Docking
Molecular docking is a computational method to predict the preferred orientation and ligand-target interactions, thus enabling scoring the molecules on how well they fit into the pocket of the target protein. Top molecules can be identified by the docking score and used in other computational methods or screened in the lab. We use ICM-Pro by MolSoft to complete the docking projects. MolSoft software performed well and outperformed a range of other methods in the numerous blinded modeling competitions and docking challenges including covalent docking, predicting protein-protein complexes [4].4D Docking (Ensemble Docking)
Using structure assembles for docking computations we can run 4D docking. This protocol works twice as fast as regular docking in the number of structures separately and opens the opportunity to consider the flexibility of the target [4, 5].RIDGE (Rapid Docking GPU Engine)
RIDGE is a fast and accurate structure-based virtual ligand screening method. Utilizing RIDGE makes it possible to dock approximately 100 compounds per second or 10M compounds database in 30h. For more details about the engine please find at Molsoft website.
3D target-performed pharmacophore modeling
By adding the excluded volumes based on information about ligand surroundings in the ligand-target complex we improve the classical pharmacophore. This approach outperforms pharmacophore screening, helps to avoid clashes, and gets more accurate results [6, 7]. To reinforce 3D pharmacophore screening as well as other ligand-based approaches, Chemspace utilizes cutting edge utilities and one of them is GINGER from Molsoft.GINGER (Fast GPU Based conformer generation)
GINGER (Graph Internal-coordinate Neural-network conformer Generator with Energy Refinement) is a new cutting-edge software for lightning-fast high-quality conformer library generation on GPUs [8]. The productivity of GINGER allows the generation of 10M compounds library in a day and quick processing of your in-house 2D combinatorial or AI-generated libraries, enabling the application of ligand 3D structure-based methods such as field-, shape- and pharmacophore-based screening. More details about GINGER please find here.