From Billions to Trillions: Addressing the Rising Complexity of Chemical Space with V-SYNTHES
Molecular docking has long been a standard technique in the virtual screening (VS) toolkit. However, its utility is increasingly challenged by the exponential growth of accessible chemical space. Over the past few years, commercially available combinatorial libraries have expanded from billions to trillions of compounds, posing a fundamental computational and methodological challenge: how to explore such massive spaces efficiently and meaningfully?
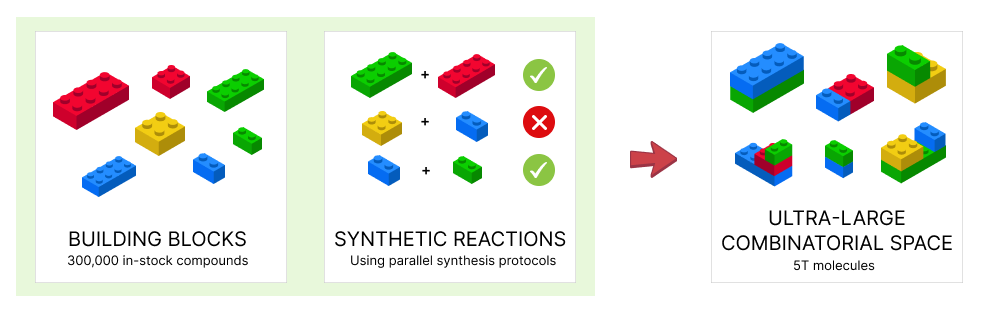
Traditional physics-based docking methods, as well as machine learning (ML)-enhanced approaches, struggle to scale with these chemical universes, especially as many of these libraries cease to exist in fully enumerated form (Luttens et al., Nat. Comput. Sci. 2025). As a result, developing methodologies capable of navigating non-enumerated chemical spaces has become critical for both early hit identification and hit-to-lead optimization.
This paradigm shift began in 2022 with the introduction of V-SYNTHES, a first-in-class method developed in the Katritch Lab (USC). V-SYNTHES demonstrated the feasibility of structure-based exploration across the (then) 11-billion-compound Enamine REAL Space, and its efficacy was validated across diverse biological targets (Sadybekov et al., Nature 2022).
Subsequently, in 2024, BioSolveIT introduced an alternative framework – Chemical Space Docking (CSD) – providing a vendor-agnostic platform for structure-guided navigation of large combinatorial libraries. Simultaneously, Molsoft, inspired by V-SYNTHES, launched CombiRIDGE, a GPU-accelerated docking system capable of screening tens of billions of compounds within days on a single workstation.
Comparative Perspectives: V-SYNTHES, CSD, and CombiRIDGE
V-SYNTHES takes full advantage of the modular nature of combinatorial spaces and does not require complete enumeration of the chemical universe. Instead, the workflow constructs a target-specific subspace based on the physicochemical properties of the binding pocket. This subspace resides entirely within the synthetically accessible Enamine REAL Space, ensuring practicality and rapid synthesis.
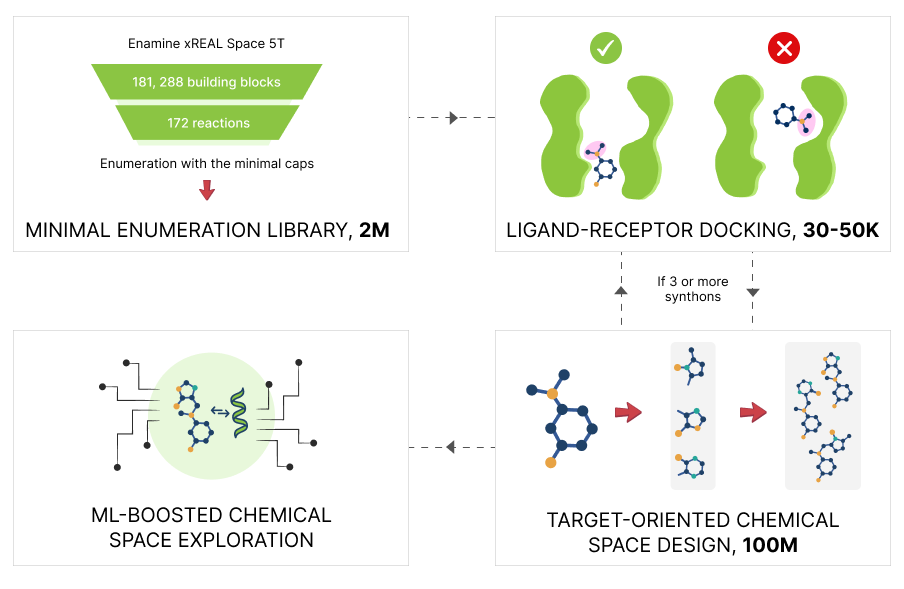
The Minimal Enumeration Library (MEL) lies at the heart of the V-SYNTHES workflow, enabling its modular and scalable design. MEL transforms chemical building blocks – specifically, synthons, digital representations of building blocks prepared for in silico assembly – into docking-ready fragments. MEL fragment-like compounds omit reactive handles and problematic groups (e.g., heavy metals like uranium or neptunium), which, if retained, could result in false positives. Importantly, MEL encodes growth vectors for downstream fragment elaboration, providing a basis for rational fragment selection.
By iteratively combining high-scoring MEL fragments and expanding only productive synthons, V-SYNTHES achieves up to 5,000-fold reductions in computational demand compared to exhaustive docking.
CSD shares conceptual similarities with V-SYNTHES, but several methodological differences exist. Notably, in CSD, the initial fragment conformations are fixed during subsequent enumeration, which may result in inaccuracies for the fully enumerated ligands. Additionally, CSD offers limited control over enumeration steps and relies heavily on visual inspection rather than classical scoring metrics, favoring qualitative over quantitative assessment. Nonetheless, its major advantage lies in integrated access to diverse vendor spaces, a user-friendly graphical user interface and the possibility to run the virtual screening behind the firewall.
By contrast, CombiRIDGE offers impressive computational performance through its a GPU-accelerated architecture, reportedly screening ~48B Enamine REAL compounds in days using a single NVIDIA RTX 4090 box. While the speed of CombiRIDGE is unprecedented, it comes with some limitations. The use of a GPU-accelerated rigid docking algorithm leads to inaccuracies, especially in shallow or exposed binding pockets. Additionally, its proprietary graph neural network (GNN)-based method for conformer generation may introduce artifacts due to incorrect handling of macrocycles or compounds with unusual motifs.
Next-Generation V-SYNTHES: CapSelect and Deep Learning Integration
Meanwhile, the Katritch Lab and Chemspace have continued to evolve the V-SYNTHES platform to keep pace with the growth of combinatorial chemical libraries, now approaching 5 trillion compounds – Enamine xREAL Space.
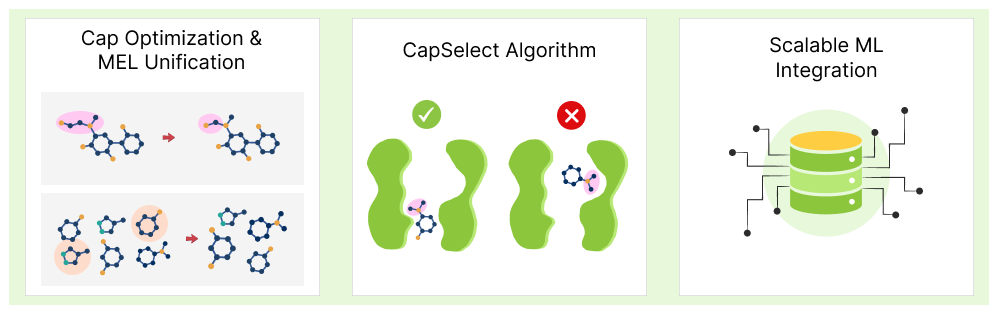
To further adapt to the leap from 11B to 5T+ compound spaces, significant enhancements have been implemented:
- Cap Optimization & MEL Unification: The selection of capping groups (used to represent generic substituents during docking) was refined to unify redundant MEL fragments across reaction types. This improves interpretability and reduces the size of the initial MEL docking set.
- CapSelect Algorithm: A new ML-driven module – CapSelect – automatically analyzes docking poses and prioritizes fragments based on their synthetically accessible growth vectors. This avoids selecting fragments sterically trapped in subpockets or capped in non-expandable orientations.
- Scalable ML Integration: To handle the combinatorial explosion (from 11B to over 5T compounds), V-SYNTHES is now coupled with deep learning and active learning modules that tailor the exploration process to each target and screening objective.
The success of the V-SYNTHES approach was first demonstrated in the original publication (Sadybekov et al., Nature, 2022), which showcased its applicability across a broad range of targets. Since then, the approach has been actively used in ongoing discovery projects in the Katritch lab, further validating its flexibility and performance. In parallel, the Chemspace team has successfully applied V-SYNTHES in real-world settings, achieving strong results in CACHE Challenges 2 and 4. Together, these efforts highlight the scalability, robustness, and broad applicability of V-SYNTHES for efficient hit identification in ultra-large chemical spaces.
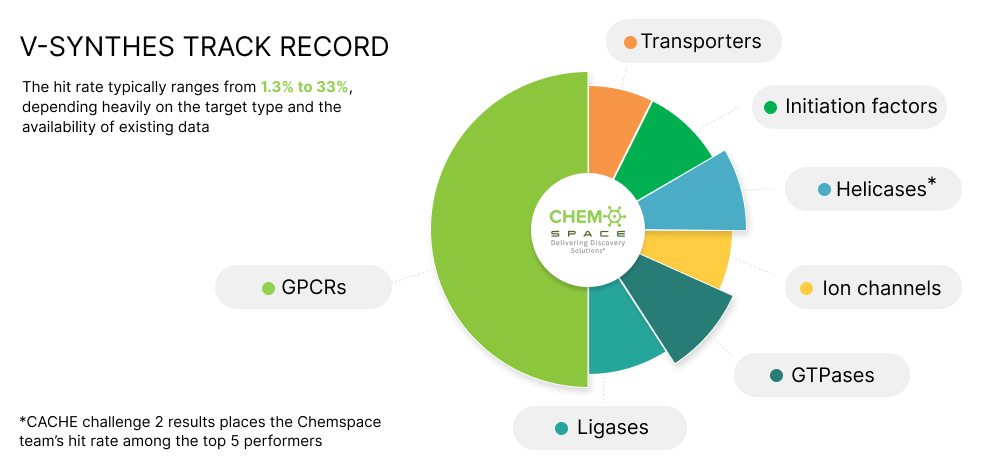
V-SYNTHES Success Stories
-
Synthon-based ligand discovery in virtual libraries of over 11 billion compounds.
Sadybekov, A.A.; Sadybekov, A.V.; Liu, Y.; Iliopoulos-Tsoutsouvas, C.; Huang, X.; Pickett, J.; Houser, B.; Patel, N.; Tran, N.K.; Tong, F.; Zvonok, N.; Jain, M.K.; Savych, O.; Radchenko, D.S.; Nikas, S.P.; Petasis, N.A.; Moroz, Y.S.; Roth, B.L.; Makriyannis, A.; Katritch, V. Nature 2021, 601 (7893), 452-459.
https://www.nature.com/articles/s41586-021-04220-9
-
CACHE Challenge #2: Targeting the RNA Site of the SARS-CoV-2 Helicase Nsp13.
Herasymenko, O.; ...; Moroz, Y.S.; ...; Protopopov, M.; ...; Tarkhanova, O.O.; ...; Schapira, M. Comput. Chem. 2025, [Online].
https://pubs.acs.org/doi/10.1021/acs.jcim.5c00535
-
Results of the CACHE Challenge #4.