Exploitation of Type II Binding Model
Type II kinase inhibitors are a distinctive class of compounds designed to selectively target kinases in their inactive state. Unlike type I inhibitors, which bind to the active form, type II inhibitors engage an extended ATP-binding site exposed through structural shifts in the kinase, revealing a hydrophobic Deep Pocket (also called the Phe or allosteric pocket). This unique binding mode allows Type II inhibitors to achieve greater specificity and affinity thereby reducing off-target effects and enhancing therapeutic precision.
Developing type II kinase inhibitors focuses on addressing drug resistance by improving binding to mutated kinases, offering new solutions in treating cancers and other kinase-driven diseases. Their distinct mechanism continues to be a promising area for advancing targeted therapies.
Library Development
Two complementary ligand-based virtual screening methods were applied to type II kinase inhibitor discovery in parallel: a fast 3D molecular similarity search method (RIDE, developed by MolSoft) and a pharmacophore-based approach.The complexes of 13 well-known inhibitors with protein kinase were extracted from PKIDB (A Curated, Annotated and Updated Database of Protein Kinase Inhibitors in Clinical Trials) and divided into 6 groups via their X-ray structure superimposition.
From these, 185 pharmacophore models were generated for virtual screening and each of the selected template compounds was used for 3D similarity search.
The conformer database was formed by generating an average of 50 conformations for each molecule from Enamine's in-stock collection (3.9M).
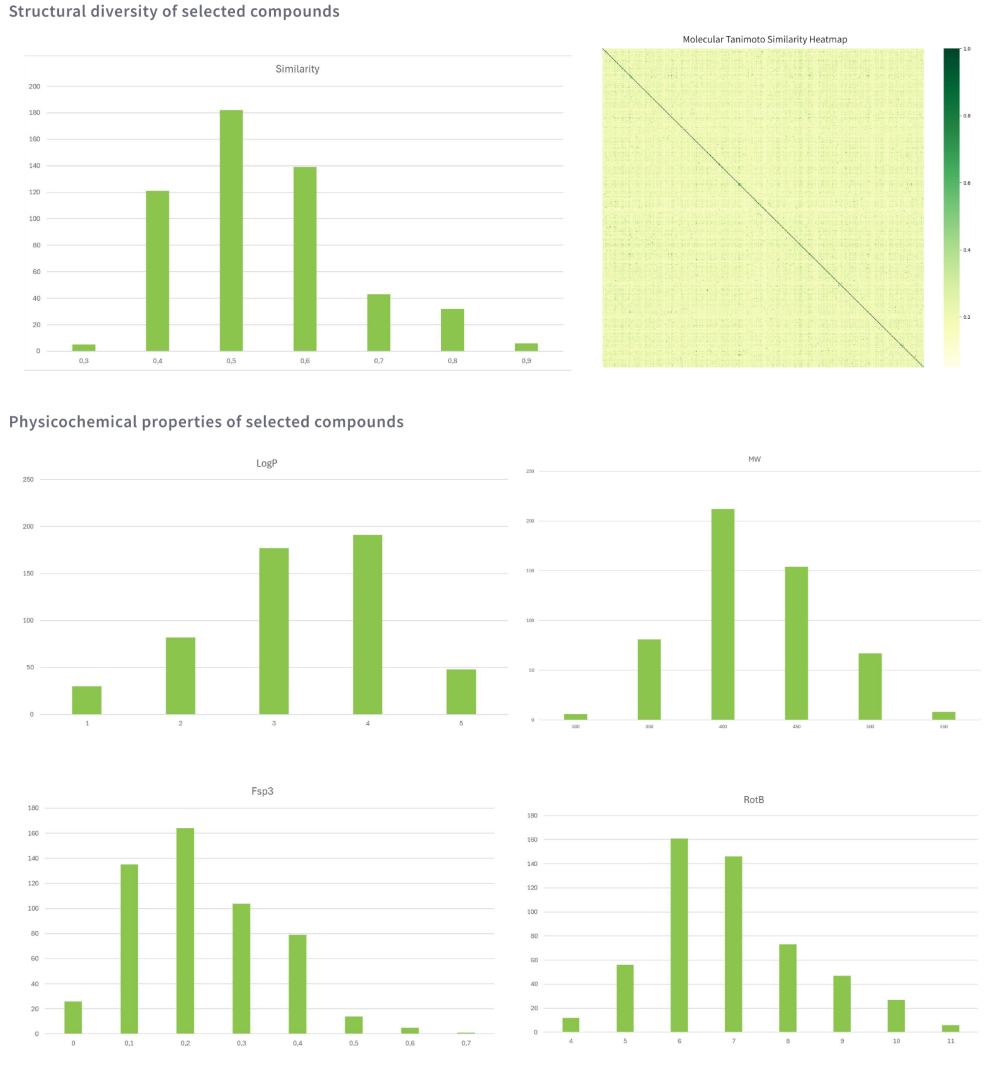
The virtual screening of the conformers database resulted in 63K and 100K perspective molecules using RIDE and the pharmacophore-based method respectively. A subset of 6K molecules that overlap between methods underwent filtering (PAINS, badgroups, ChEMBL similarity (≤ 0.7)) and expert evaluation. Finally, the library of 528 potential type II kinase inhibitors was pre-plated.
Please refer to the attached PDF file for more detailed information about this process.
Our Offering
Following the screening of 3.9M compounds, we curated a library of 528 potent and novel type II kinase inhibitors.The graphs below showcase the key physicochemical properties of compounds.
Download the library now and start exploring its potential!
Should you require any additional information, feel free to contact us at [email protected], and we will gladly fulfill your request!