“Crystal Structure First” Approach
Fragment-Based Drug Discovery (FBDD) has proven itself as a reliable method to discover starting points that can be optimized to become clinical candidates. After the identification of potent fragments, the fragment growth stage becomes a challenge due to the cost and time needed to synthesize custom compounds. The “Crystal Structure First” Approach brings the FBDD to a new level by utilizing the chemical modularity of Enamine REAL Space. Using this approach, we at Chemspace perform the fragment evolution to the full-size molecule hits with a remarkable boost in potency and at the price of REAL compounds. [1]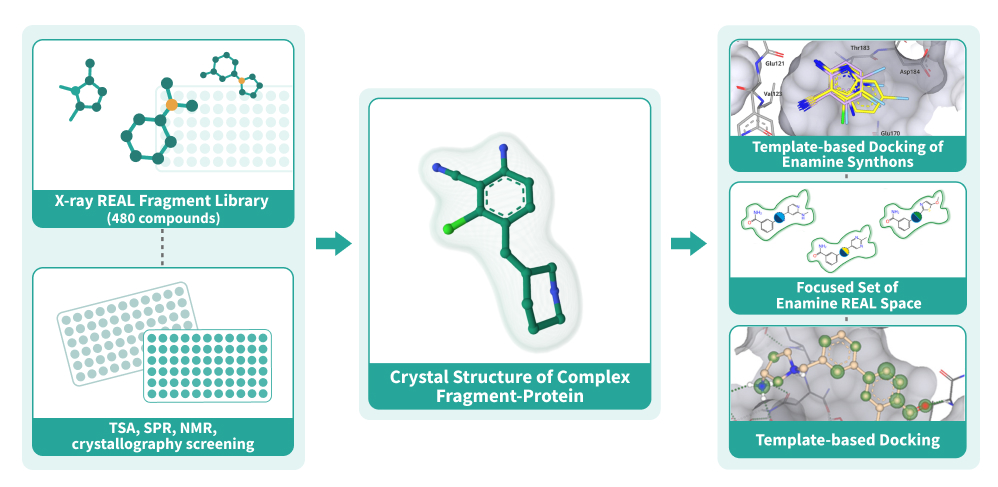
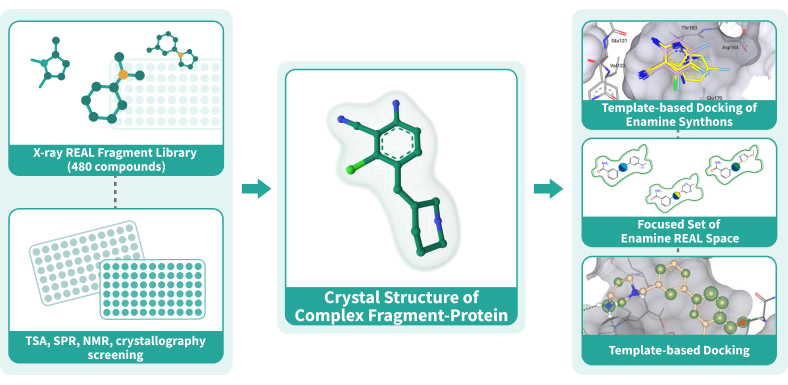
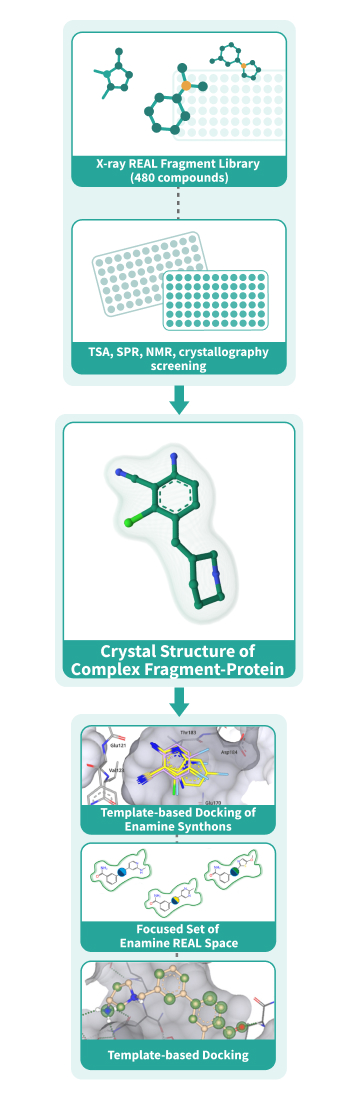 Utilizing the power of our fragment libraries, we can detect the fragment(s) that bind to the target,
crystalize the fragment-target complexes, and use this information to perform template-based docking
of Enamine REAL Space. The molecular docking is done using the state-of-the-art software ICM-Pro by
MolSoft.
Utilizing the power of our fragment libraries, we can detect the fragment(s) that bind to the target,
crystalize the fragment-target complexes, and use this information to perform template-based docking
of Enamine REAL Space. The molecular docking is done using the state-of-the-art software ICM-Pro by
MolSoft.
REAL crystallographic Fragment Library, 480 compounds
Screening the fragments using crystallography results in detection of binding modes right after the first screen but has a much lower throughput. This is why we created a library of 480 compounds based on the REAL Fragment Library for you to be able to benefit from this approach. The compounds were selected using fingerprint-based similarity to ensure their diversity, so we can provide maximum chemical space coverage with less compounds.Chemical space
Crystal Structure First can be applied to Enamine xREAL Space of 2.3 trillion molecules.Project requirements
The workflow requires crystal structure(s) of the fragment(s) bound to the target. You can provide the structures from your in-house screens, or we perform a pre-selection stage to identify the initial fragments together. Additional requirements for hit selection can be discussed upon request.Workflow description
There are two ways of entering the “Crystal Structure First” workflow:- Pre-selection stage. For identification of initial fragments, we provide REAL Fragment Library and REAL Crystallographic Fragment Library, but other fragment libraries can be used as well. REAL Fragment Library can be screened by TSA or SPR methods. The potential binders will be crystalized to get the binding modes of the fragments. Using crystallography to screen the REAL Crystallographic Fragment Library we can get the binders and the binding modes for them after just one screen.
- Template-based docking stage. If you have already completed a fragment screen, we use the obtained information to set up template-based molecular docking of the Enamine Synthons to select the synthons for enumeration. The main criteria for synthon prioritization are docking score, APF score, the direction of the reaction vectors, and ligand efficiency.
References
- Müller, J.; Klein, R.; Tarkhanova, O.; Gryniukova, A.; Borysko, P.; Merkl, S.; Ruf, M.; Neumann, A.; Gastreich, M.; Moroz, Y. S.; Klebe, G.; Glinca, S. Magnet for the Needle in Haystack: “Crystal Structure First” Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces. J. Med. Chem. 2022, 65, 23, 15663–15678. https://doi.org/10.1021/acs.jmedchem.2c00813 .